 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Ote100226460061 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lamiaceae; Nepetoideae; Ocimeae; Ocimum
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 424aa MW: 48212.2 Da PI: 5.0829 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 79.4 | 5.3e-25 | 56 | 136 | 2 | 86 |
trihelix 2 WtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyf 82
W ++e++ Li +r+e++ ++++k++k+lWe +s kmre+gf rsp++C++kw+nl k+ykk+k+++++++ +++++
Ote100226460061|100226460061 56 WAQEETRSLICLRKEIDFLFNTSKSNKHLWENISMKMREKGFDRSPTMCTDKWRNLLKEYKKTKQNNQDGN----AKMSFY 132
********************************************************************983....357799 PP
trihelix 83 dqle 86
+++e
Ote100226460061|100226460061 133 KDIE 136
9987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-4 | 47 | 117 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50090 | 8.386 | 48 | 112 | IPR017877 | Myb-like domain |
| SMART | SM00717 | 5.9E-5 | 52 | 114 | IPR001005 | SANT/Myb domain |
| CDD | cd12203 | 1.88E-26 | 54 | 119 | No hit | No description |
| Pfam | PF13837 | 1.2E-19 | 55 | 138 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 424 aa Download sequence Send to blast |
MLMPEKAQEQ EETAIEFYST KEASPNGNNM IIQIGDGDDQ NPPPPSPAPK KRAETWAQEE 60 TRSLICLRKE IDFLFNTSKS NKHLWENISM KMREKGFDRS PTMCTDKWRN LLKEYKKTKQ 120 NNQDGNAKMS FYKDIEEIMS ERGKNGHCWK NSVASSDKVD SFMQFSDKVV EIGVGFLYSS 180 VGXXXXXXXX ASKSFIAGKR LVEKCSVLHD CLILFGSLLL FFCEVAQSGI DDASLTFGPV 240 EDNGRSTLNL ERPLDHEAHP LAITAADASG VSPWNWRETP GNGENRSNYD GRVITVKLGD 300 YTKRIGIDGT ADAIKEALES AFRLRTKRAF WLEDEDNVVR SLDRDMPLGN YTLHVDEGLT 360 IKICLFDGSE HLPVHTEEKT FYTEDDFRGF LSRRCWTYLR EFDGYRNVDS MDELCPGALY 420 RGVN |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2jmw_A | 7e-38 | 50 | 134 | 1 | 86 | DNA binding protein GT-1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
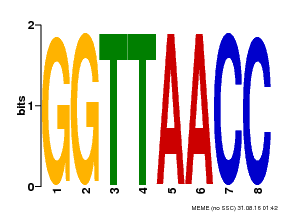
| UniProt | Probable transcription factor that binds specifically to the core DNA sequence 5'-GGTTAA-3'. May act as a molecular switch in response to light signals. {ECO:0000269|PubMed:10437822, ECO:0000269|PubMed:15044016, ECO:0000269|PubMed:7866025}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00004 | PBM | Transfer from 920224 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011071308.1 | 0.0 | trihelix transcription factor GT-4-like | ||||
| Swissprot | Q9FX53 | 1e-137 | TGT1_ARATH; Trihelix transcription factor GT-1 | ||||
| TrEMBL | A0A4D9B8M9 | 0.0 | A0A4D9B8M9_SALSN; Uncharacterized protein | ||||
| STRING | Migut.E00236.1.p | 1e-170 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA5113 | 21 | 36 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



