 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Ote100248320071 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lamiaceae; Nepetoideae; Ocimeae; Ocimum
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 457aa MW: 52768.6 Da PI: 7.6214 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 11.1 | 0.0012 | 19 | 41 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
y C++Cg + s k + +Hir H
Ote100248320071|100248320071 19 YYCEYCGICRSKKALIASHIRAH 41
89*******************96 PP
| |||||||
| 2 | zf-C2H2 | 21.6 | 5.7e-07 | 63 | 84 | 2 | 23 |
EETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 2 kCpdCgksFsrksnLkrHirtH 23
+C+ Cg sF+ + +Lk+H+++H
Ote100248320071|100248320071 63 VCEECGASFRKPAHLKQHMQSH 84
6*******************99 PP
| |||||||
| 3 | zf-C2H2 | 17.7 | 9.6e-06 | 127 | 151 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
+ C+ dC s++rk++L+rH+ H
Ote100248320071|100248320071 127 FICSvdDCQSSYRRKDHLTRHMLQH 151
789999****************988 PP
| |||||||
| 4 | zf-C2H2 | 18.8 | 4.6e-06 | 156 | 181 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
++Cp dC+++F+ +sn +rH + H
Ote100248320071|100248320071 156 FECPveDCKRTFTLQSNVTRHVKGfH 181
89*******************99777 PP
| |||||||
| 5 | zf-C2H2 | 14.6 | 9.7e-05 | 212 | 236 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirt.H 23
+Cp dC+++F++ ++k H + H
Ote100248320071|100248320071 212 ECPvqDCKRTFKSEGHMKWHVKEfH 236
69999****************9988 PP
| |||||||
| 6 | zf-C2H2 | 16.8 | 1.8e-05 | 250 | 274 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
++C Cgk+F+ +s L +H +H
Ote100248320071|100248320071 250 HVCAdpQCGKVFKYPSKLSKHEDSH 274
688888***************9888 PP
| |||||||
| 7 | zf-C2H2 | 20.1 | 1.7e-06 | 341 | 365 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirt.H 23
+C+ C+++Fs+ snL++Hi+ H
Ote100248320071|100248320071 341 SCSvkGCSRTFSTTSNLNQHIKAvH 365
588889***************9888 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 1.65E-9 | 17 | 84 | No hit | No description |
| PROSITE profile | PS50157 | 9.245 | 19 | 46 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 21 | 19 | 41 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 7.9E-7 | 58 | 85 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 12.03 | 62 | 89 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0026 | 62 | 84 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 64 | 84 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 8.9E-10 | 124 | 153 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 6.72E-7 | 124 | 153 | No hit | No description |
| SMART | SM00355 | 0.0034 | 127 | 151 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 9.723 | 127 | 156 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 129 | 151 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 3.2E-5 | 154 | 179 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 3.86E-5 | 154 | 182 | No hit | No description |
| PROSITE profile | PS50157 | 10.284 | 156 | 181 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.013 | 156 | 181 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 158 | 181 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0058 | 211 | 236 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 8.912 | 211 | 237 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 213 | 236 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 4.3E-4 | 250 | 274 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.2 | 250 | 274 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.45 | 250 | 279 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 252 | 274 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 2.4 | 282 | 307 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 284 | 307 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 13 | 310 | 331 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 6.3E-7 | 321 | 362 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 6.83E-10 | 321 | 381 | No hit | No description |
| PROSITE profile | PS50157 | 12.05 | 340 | 370 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0014 | 340 | 365 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 342 | 365 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 9.4E-6 | 369 | 393 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 6.3 | 371 | 397 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 373 | 397 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 457 aa Download sequence Send to blast |
MVDEGEKPRA PIFRDIRRYY CEYCGICRSK KALIASHIRA HHEEELKEKE LNEADEHGEK 60 LNVCEECGAS FRKPAHLKQH MQSHSLENSK MREAAILLDW ISTRWVTAWE APPSLPRSRL 120 LNFERPFICS VDDCQSSYRR KDHLTRHMLQ HEGKLFECPV EDCKRTFTLQ SNVTRHVKGF 180 HQESACANLE HPDDSDSLES DKHEDSSGKL LECPVQDCKR TFKSEGHMKW HVKEFHSESA 240 CADVNHPLGH VCADPQCGKV FKYPSKLSKH EDSHVKLDTV EAFCAEPGCM KYFSNEQCLK 300 EHIRSCHQYV VCDECGTKHL KKNIKRHMRS HEGELTPGRI SCSVKGCSRT FSTTSNLNQH 360 IKAVHWELKP FACGIPGCDK RFTYKHVRDN HEKSGCHIYT AGDFGASDEQ FRLRPRGGRK 420 RTYPVIETLM RKRIVPPTES ELVGNEGSWQ IYSEYED |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Essential protein (PubMed:22353599). Isoform 1 is a transcription activator the binds both 5S rDNA and 5S rRNA and stimulates the transcription of 5S rRNA gene (PubMed:12711688, PubMed:22353599). Isoform 1 regulates 5S rRNA levels during development (PubMed:22353599). {ECO:0000269|PubMed:12711688, ECO:0000269|PubMed:22353599}. | |||||
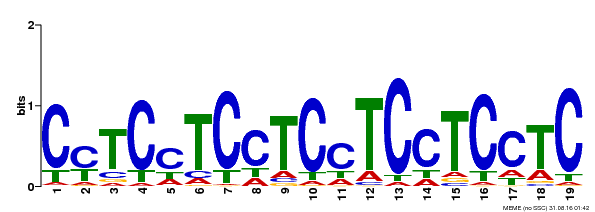
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011079244.1 | 1e-174 | transcription factor IIIA | ||||
| Swissprot | Q84MZ4 | 1e-139 | TF3A_ARATH; Transcription factor IIIA | ||||
| TrEMBL | A0A4D8Z9A3 | 0.0 | A0A4D8Z9A3_SALSN; Uncharacterized protein | ||||
| STRING | Migut.K00168.1.p | 1e-164 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3644 | 24 | 47 |



