 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Ote100267110011 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lamiaceae; Nepetoideae; Ocimeae; Ocimum
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 302aa MW: 33839.4 Da PI: 4.6338 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 59.6 | 5e-19 | 59 | 112 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k+++++ +q+++Le+ Fe ++++ e++ +LA++lgL+ rqV vWFqNrRa++k
Ote100267110011|100267110011 59 KKRRLSVDQVKALEKNFEVENKLEPERKVKLAQELGLQPRQVAVWFQNRRARWK 112
566899***********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 129.5 | 1.3e-41 | 58 | 150 | 1 | 93 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLe 81
ekkrrls +qvk+LE++Fe e+kLeperKv+la+eLglqprqvavWFqnrRAR+ktkqlE+dy +Lk+++dalk + e+L+
Ote100267110011|100267110011 58 EKKRRLSVDQVKALEKNFEVENKLEPERKVKLAQELGLQPRQVAVWFQNRRARWKTKQLERDYGVLKANHDALKLKYETLQ 138
69******************************************************************************* PP
HD-ZIP_I/II 82 keveeLreelke 93
+e+e+L++e++e
Ote100267110011|100267110011 139 RENESLHNEIRE 150
********9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.87E-19 | 48 | 116 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.119 | 54 | 114 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 6.3E-18 | 57 | 118 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.39E-17 | 59 | 115 | No hit | No description |
| Pfam | PF00046 | 3.2E-16 | 59 | 112 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 5.9E-20 | 61 | 121 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 8.3E-6 | 85 | 94 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 89 | 112 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 8.3E-6 | 94 | 110 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 4.0E-16 | 114 | 155 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009637 | Biological Process | response to blue light | ||||
| GO:0009788 | Biological Process | negative regulation of abscisic acid-activated signaling pathway | ||||
| GO:0030308 | Biological Process | negative regulation of cell growth | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048510 | Biological Process | regulation of timing of transition from vegetative to reproductive phase | ||||
| GO:0048573 | Biological Process | photoperiodism, flowering | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 302 aa Download sequence Send to blast |
MKRLSSSDSL AGLMSICPNS TVDEHNHGNN PIYTRDFQTM MDGLEEEGCV EESCNISEKK 60 RRLSVDQVKA LEKNFEVENK LEPERKVKLA QELGLQPRQV AVWFQNRRAR WKTKQLERDY 120 GVLKANHDAL KLKYETLQRE NESLHNEIRE LKLKLNGETS VKEEASLSEN KEKSDVEIHS 180 GGAAAELNPE NCDAKENETV AIFGDFKDGS SDSDSSVILN EDNNSPIAAA ISSSGGILHH 240 RFSDIDGGGG LTNAAVNAKF GFFDAPLVKI EEHNFFGEES CTTLFSDDQA PTLHWYCNDE 300 WN |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00271 | DAP | Transfer from AT2G22430 | Download |
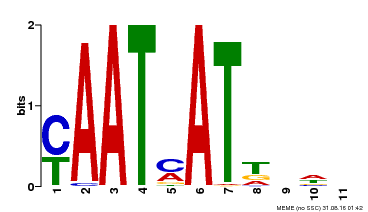
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011097532.1 | 1e-119 | homeobox-leucine zipper protein ATHB-6 | ||||
| TrEMBL | A0A2G9I4D6 | 1e-123 | A0A2G9I4D6_9LAMI; Transcription factor HEX | ||||
| STRING | Migut.L00161.1.p | 1e-112 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA2718 | 24 | 53 |



