 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Ote100268020101 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lamiaceae; Nepetoideae; Ocimeae; Ocimum
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 578aa MW: 65290.5 Da PI: 5.1719 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 49.2 | 2.1e-15 | 1 | 106 | 47 | 131 |
DUF822 47 nneVlkALcreAGwvvedDGttyr...................kgskpl.eeaeaagssasaspesslq.sslkssalasp 106
+n+V++AL+reAGw+ve+DGttyr ++ +++ ++a+aag +s+es+l s+l++++++s
Ote100268020101|100268020101 1 MNDVIAALAREAGWTVEPDGTTYRqtspaplqpqsqplpishfNSIQQNnSNASAAGPNPVRSVESPLYsSALRNCSVRSS 81
7***********************444433333222222222222222222223333333344444444344555555555 PP
DUF822 107 vesysaspksssfpspssldsisla 131
v+ + ++++ ++ +sp+slds+ ++
Ote100268020101|100268020101 82 VDCQPSTLRVDENMSPASLDSVVAE 106
5555555555555555555555444 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 1.5E-17 | 1 | 122 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| SuperFamily | SSF51445 | 9.0E-143 | 147 | 575 | IPR017853 | Glycoside hydrolase superfamily |
| Gene3D | G3DSA:3.20.20.80 | 2.1E-154 | 147 | 575 | IPR013781 | Glycoside hydrolase, catalytic domain |
| Pfam | PF01373 | 9.8E-72 | 172 | 555 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.4E-48 | 205 | 223 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.4E-48 | 227 | 248 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.4E-48 | 320 | 342 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.4E-48 | 393 | 412 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.4E-48 | 427 | 443 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.4E-48 | 444 | 455 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.4E-48 | 462 | 485 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.4E-48 | 500 | 522 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 578 aa Download sequence Send to blast |
MNDVIAALAR EAGWTVEPDG TTYRQTSPAP LQPQSQPLPI SHFNSIQQNN SNASAAGPNP 60 VRSVESPLYS SALRNCSVRS SVDCQPSTLR VDENMSPASL DSVVAERDAK LDKYSSASAI 120 NSPECLEPSQ LVQDVHCREH GNGLPETSYV PVYVRLNTGL INNFCQLMDP EGVKEELQHL 180 RSLXXXXXXX XXXXGIVEAW TPKKYEWAGY REIFNIIREF GMKLQVVMAF HEYGGTDASG 240 IFVSLPQWVL EIGKSNQDIF FTDREGRRNT ECLSWGVDKE RVLKGRTGIE IYFDFMRSFR 300 TEFDDLFTEG LISAVEVGLG ASGELKYPSF SERLGWRYPG IGEFQCYDKY LLQNLQKAAK 360 LRGHSFWARG PDNAGFYNSK PHETGFFCDR GDYDSYYGRF FLQWYNQVLI DHADNVLSLA 420 TLAFEDIKIV VKIPAVYWWY KTASHAAELT AGYYNSSNHD GYALLFEVLK KHSVAMKFVL 480 SGFQTSYPEI NEALSDPEGL NWQVLNCGWD RGLSVGGENA EPCYNRDGLM RLVEAAKPRD 540 HPDHLHFSFL VFQQPIQTSV CFSEIDHFVK SMHGELSN |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2dqx_A | 1e-116 | 148 | 575 | 10 | 444 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
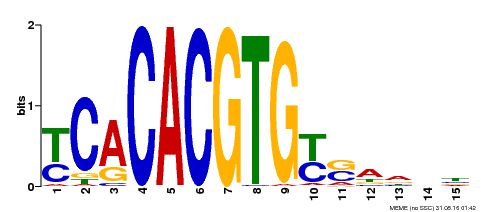
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011082155.1 | 0.0 | beta-amylase 8 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | A0A4D9BKE8 | 0.0 | A0A4D9BKE8_SALSN; Uncharacterized protein | ||||
| STRING | Migut.A00486.1.p | 0.0 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA1607 | 24 | 48 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



