 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Ote100268410001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lamiaceae; Nepetoideae; Ocimeae; Ocimum
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 567aa MW: 62659.6 Da PI: 7.9846 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 12.2 | 0.00033 | 263 | 280 | 2 | 19 |
HHHHHHHHHHHHHHHHHH CS
HLH 2 rrahnerErrRRdriNsa 19
r +h++ E+rRR++iN++
Ote100268410001|100268410001 263 RSKHSATEQRRRSKINDR 280
889*************96 PP
| |||||||
| 2 | HLH | 25.1 | 3.1e-08 | 323 | 355 | 19 | 55 |
HHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 19 afeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
f++Lre++P++ ++K +Ka+ L +++eYI+ Lq
Ote100268410001|100268410001 323 LFHKLREIIPNS----DQKRDKASFLLEVIEYIQFLQ 355
699********9....9*******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 3.1E-18 | 263 | 280 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 7.10E-8 | 263 | 358 | No hit | No description |
| Pfam | PF00010 | 1.4E-5 | 323 | 355 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 9.55E-10 | 323 | 382 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 3.1E-18 | 324 | 377 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0009742 | Biological Process | brassinosteroid mediated signaling pathway | ||||
| GO:1902448 | Biological Process | positive regulation of shade avoidance | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 567 aa Download sequence Send to blast |
MELPFGTEGR KATHDFLSLY SSPAAHTQQD PSPQQGGYLK THDFLQPLER EGKNVTTTKE 60 DIKAGLAAVD RPSVPAPPAS VEHILPGGIG TYSISYFNQM ALKPEGSLFT AQPSSTARND 120 ENSNCTSYPG GSFTLWDESA IKKGKTGKEN ISVERHILRE AGVNVVGGQW TSFLEKPSRS 180 SSNHRHNATN FSSLSSSQPS SSQKNQSFMD MMMTSTKNDL XXXXXXXXXX FMSIKKEPSC 240 HLKGNLSVKV EAKTIDQKPN TPRSKHSATE QRRRSKINDR HVYQPPQFEV QDFDQLSEFA 300 SVFPCYIMSS YHHPESFLGL FTLFHKLREI IPNSDQKRDK ASFLLEVIEY IQFLQEKVNR 360 YENSYNVWNQ ESSKMMHWQR NCHINEGVID RTVGVSSDSA PTALVFGKKF EENKGGVSPS 420 LPTSGQNLVD SDLSSDTTLR ERVQPPILTT KGLANILSLG SISVPPKLGS EVDKSTTRPQ 480 SQLLRSCNKM TEQEVSIESG TINISSVYSQ GLLNTLTQAL ESSGVDLSQA SVSVQIDLGK 540 RASSLVSKVD EGGKEELGHA SKKVRSS |
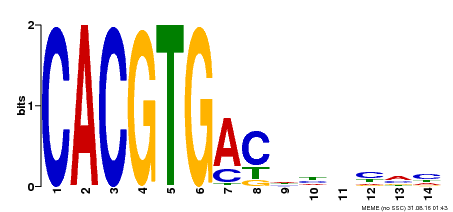
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00498 | DAP | Transfer from AT5G08130 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011078682.1 | 0.0 | transcription factor BIM1 isoform X1 | ||||
| TrEMBL | A0A4D8ZFI0 | 0.0 | A0A4D8ZFI0_SALSN; Uncharacterized protein | ||||
| STRING | Migut.D01611.1.p | 0.0 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3969 | 23 | 45 |



