 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PDK_30s1016001g009 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Coryphoideae; Phoeniceae; Phoenix
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 334aa MW: 38633.3 Da PI: 7.0013 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 182 | 1.5e-56 | 7 | 136 | 1 | 129 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpk..kvka.eekewyfFskrdkkyatgkrknratksgyWkatgk 88
+ppGfrFhPtdeelv++yL+kkv+++ ++l +vik+vd+yk+ePwdL++ k+ + e++ewyfFs++dkky+tg+r+nrat++g+Wkatg+
PDK_30s1016001g009 7 VPPGFRFHPTDEELVDYYLRKKVASRGIDL-DVIKDVDLYKIEPWDLQEkcKIGTeEQNEWYFFSHKDKKYPTGTRTNRATAAGFWKATGR 96
69****************************.9***************952444443555******************************** PP
NAM 89 dkevlskkgelvglkktLvfykgrapkgektdWvmheyrle 129
dk+++s k++l+g++ktLvfykgrap+g+k+dW+mheyrle
PDK_30s1016001g009 97 DKPIYS-KNNLIGMRKTLVFYKGRAPNGQKSDWIMHEYRLE 136
******.899*****************************85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 3.27E-62 | 4 | 156 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 60.773 | 7 | 156 | IPR003441 | NAC domain |
| Pfam | PF02365 | 6.7E-29 | 8 | 135 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048759 | Biological Process | xylem vessel member cell differentiation | ||||
| GO:1901348 | Biological Process | positive regulation of secondary cell wall biogenesis | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 334 aa Download sequence Send to blast |
MTTFSHVPPG FRFHPTDEEL VDYYLRKKVA SRGIDLDVIK DVDLYKIEPW DLQEKCKIGT 60 EEQNEWYFFS HKDKKYPTGT RTNRATAAGF WKATGRDKPI YSKNNLIGMR KTLVFYKGRA 120 PNGQKSDWIM HEYRLETNEN GTPQEEGWVV CRVFKKRVTA MRGVSEHESP RWYDEQSSFM 180 QDLDSAKQIA PRPDMAYHHH LYSCKPEQDL PHLMPNDSFL QLPQLESPKL PNNVNHACDL 240 QSSTIRPEEP MQHGRQLQII SIYNSSGSTD QSAEQVTDWR VLDKFVASQL SHDVVSKEPN 300 YSNPAQVFHL SDKKEATVEY ASTSTSSGQI NLWK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 4e-52 | 7 | 162 | 17 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 4e-52 | 7 | 162 | 17 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 4e-52 | 7 | 162 | 17 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 4e-52 | 7 | 162 | 17 | 171 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 4e-52 | 7 | 162 | 20 | 174 | NAC domain-containing protein 19 |
| 3swm_B | 4e-52 | 7 | 162 | 20 | 174 | NAC domain-containing protein 19 |
| 3swm_C | 4e-52 | 7 | 162 | 20 | 174 | NAC domain-containing protein 19 |
| 3swm_D | 4e-52 | 7 | 162 | 20 | 174 | NAC domain-containing protein 19 |
| 3swp_A | 4e-52 | 7 | 162 | 20 | 174 | NAC domain-containing protein 19 |
| 3swp_B | 4e-52 | 7 | 162 | 20 | 174 | NAC domain-containing protein 19 |
| 3swp_C | 4e-52 | 7 | 162 | 20 | 174 | NAC domain-containing protein 19 |
| 3swp_D | 4e-52 | 7 | 162 | 20 | 174 | NAC domain-containing protein 19 |
| 4dul_A | 4e-52 | 7 | 162 | 17 | 171 | NAC domain-containing protein 19 |
| 4dul_B | 4e-52 | 7 | 162 | 17 | 171 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the secondary wall NAC binding element (SNBE), 5'-(T/A)NN(C/T)(T/C/G)TNNNNNNNA(A/C)GN(A/C/T)(A/T)-3', in the promoter of target genes (By similarity). Involved in xylem formation by promoting the expression of secondary wall-associated transcription factors and of genes involved in secondary wall biosynthesis and programmed cell death, genes driven by the secondary wall NAC binding element (SNBE). Triggers thickening of secondary walls (PubMed:16103214, PubMed:25148240). {ECO:0000250|UniProtKB:Q9LVA1, ECO:0000269|PubMed:16103214, ECO:0000269|PubMed:25148240}. | |||||
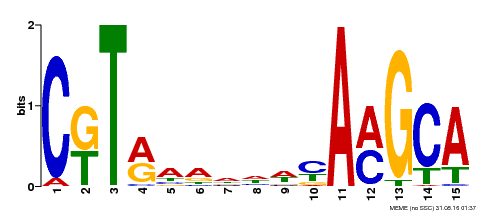
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00136 | DAP | Transfer from AT1G12260 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008796844.1 | 0.0 | NAC domain-containing protein 7 | ||||
| Swissprot | Q9FWX2 | 1e-135 | NAC7_ARATH; NAC domain-containing protein 7 | ||||
| TrEMBL | A0A2H3YCP6 | 0.0 | A0A2H3YCP6_PHODC; NAC domain-containing protein 7 | ||||
| STRING | XP_008796844.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2370 | 38 | 92 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G12260.1 | 1e-127 | NAC 007 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



