 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PDK_30s1033551g001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Coryphoideae; Phoeniceae; Phoenix
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 332aa MW: 37290.9 Da PI: 8.4945 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 34.8 | 3.7e-11 | 44 | 77 | 3 | 36 |
XXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 3 elkrerrkqkNReAArrsRqRKkaeieeLeekvk 36
+ k rr+++NReAAr+sR+RKka++++Le
PDK_30s1033551g001 44 DAKTLRRLAQNREAARKSRLRKKAYVQQLESSKM 77
67999************************97443 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 1.3E-8 | 37 | 89 | No hit | No description |
| SMART | SM00338 | 4.3E-8 | 42 | 124 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.163 | 44 | 88 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 2.3E-8 | 45 | 82 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 9.14E-7 | 46 | 87 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 49 | 64 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF14144 | 2.4E-31 | 133 | 207 | IPR025422 | Transcription factor TGA like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 332 aa Download sequence Send to blast |
MELPNPRGDL AALTEPQKDI KAVVKREGNG KGPSSEQEEP KTPDAKTLRR LAQNREAARK 60 SRLRKKAYVQ QLESSKMRLA QLEHELQRAR AQGFLVGSGS LPAGSTGGGG LPDDAAMFDV 120 EYGKWLVDHH RLMCEMRAAM QEHLPENELR MLVDKCLLHY DAVMSLKSVM ARSDAFHLYS 180 GMWMTPAERC FMWIGGFRPS ELIKIVLRHI EPLTEQQIVG VCGLQQSVHE TEEALSQGLE 240 ALFRSLSDTI ISDALGSPSD VGNYISQMVV AMNKLSALEA FVRQADNLRQ QTVHRLQLIL 300 STHQAARCFL AMAEHFHRLR ALSSLWLTRP RQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to as1-like elements (5'-TGACGTAAgggaTGACGCA-3') in promoters of target genes. Regulates transcription in response to plant signaling molecules salicylic acid (SA), methyl jasmonate (MJ) and auxin (2,4D) only in leaves. Prevents lateral branching and may repress defense signaling. {ECO:0000269|PubMed:12777042}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00491 | DAP | Transfer from AT5G06839 | Download |
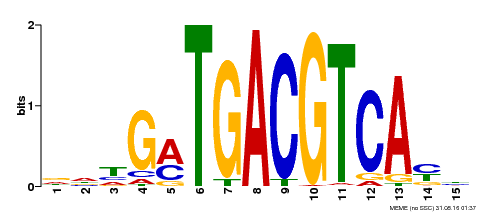
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010928841.1 | 0.0 | bZIP transcription factor TGA10 isoform X1 | ||||
| Refseq | XP_029122052.1 | 0.0 | bZIP transcription factor TGA10 isoform X2 | ||||
| Swissprot | Q52MZ2 | 1e-163 | TGA10_TOBAC; bZIP transcription factor TGA10 | ||||
| TrEMBL | A0A2H3ZN24 | 1e-178 | A0A2H3ZN24_PHODC; bZIP transcription factor TGA10-like isoform X3 | ||||
| STRING | XP_008783086.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP7876 | 35 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G06839.3 | 1e-134 | bZIP family protein | ||||



