 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PDK_30s1086741g004 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Coryphoideae; Phoeniceae; Phoenix
|
||||||||
| Family | RAV | ||||||||
| Protein Properties | Length: 306aa MW: 34028.6 Da PI: 10.3564 | ||||||||
| Description | RAV family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 31.1 | 6e-10 | 24 | 65 | 1 | 48 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaia 48
s+ykGV + +grW A+I++ ++r++lg+f + +Aa+a++a
PDK_30s1086741g004 24 SRYKGVVPQP-NGRWGAQIYEK-----HQRVWLGTFNEESQAARAYEA 65
78****9888.8*********3.....5**********88******87 PP
| |||||||
| 2 | B3 | 97.2 | 1.1e-30 | 138 | 237 | 1 | 96 |
EEEE-..-HHHHTT-EE--HHH.HTT....---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SS CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh....ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgr 87
f k tpsdv+k++rlv+pk++ae+h g+ ++++ l +ed sg+ W+++++y+++s++yvltkGW++Fvk++ Lk+gD+v F +++
PDK_30s1086741g004 138 FSKPVTPSDVGKLNRLVIPKQHAEKHfplkFGSATTGMLLNFEDVSGKIWRFRYSYWNSSQSYVLTKGWSRFVKEKSLKAGDVVGFWRSRG 228
7899**************************6677899*************************************************98877 PP
SEE..EEEE CS
B3 88 sefelvvkv 96
e++l++ +
PDK_30s1086741g004 229 PEKQLYIDW 237
777777766 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 1.18E-10 | 24 | 66 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 9.8E-6 | 24 | 65 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 4.45E-15 | 24 | 69 | No hit | No description |
| PROSITE profile | PS51032 | 14.157 | 25 | 65 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 6.9E-13 | 25 | 65 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 4.0E-15 | 25 | 76 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:2.40.330.10 | 2.2E-39 | 136 | 240 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 8.5E-31 | 137 | 238 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.04E-29 | 137 | 231 | No hit | No description |
| PROSITE profile | PS50863 | 13.38 | 138 | 239 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 2.6E-27 | 138 | 237 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 7.6E-26 | 138 | 241 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 306 aa Download sequence Send to blast |
MGSVVLDPTP DMGFETESRK LPSSRYKGVV PQPNGRWGAQ IYEKHQRVWL GTFNEESQAA 60 RAYEAVTNFT PLSETDDDGA AELAFLASHS KAEIVDMMRK HTYCDELRQS KRAGKASRGT 120 VGKRNPPSSL GLARDLLFSK PVTPSDVGKL NRLVIPKQHA EKHFPLKFGS ATTGMLLNFE 180 DVSGKIWRFR YSYWNSSQSY VLTKGWSRFV KEKSLKAGDV VGFWRSRGPE KQLYIDWKPR 240 GVAADATVPR AGHLQVVRLF GVNIFRTPVG CGGGNGSGGD GKSGGERELF PPEQFVKKQR 300 VGALWY |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wid_A | 5e-53 | 135 | 252 | 11 | 130 | DNA-binding protein RAV1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). Transcriptional repressor of flowering time on long day plants. Acts directly on FT expression by binding 5'-CAACA-3' and 5'-CACCTG-3 sequences (Probable). Functionally redundant with TEM1. {ECO:0000250, ECO:0000269|PubMed:18718758, ECO:0000305}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00025 | PBM | Transfer from AT1G68840 | Download |
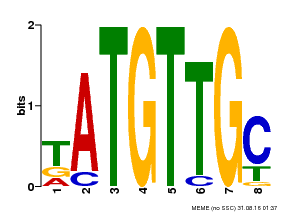
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | JX997989 | 1e-39 | JX997989.1 Antirrhinum majus isolate B10.8.2.1 tempranillo (TEM) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017696993.1 | 0.0 | AP2/ERF and B3 domain-containing transcription repressor RAV2-like | ||||
| Swissprot | P82280 | 1e-114 | RAV2_ARATH; AP2/ERF and B3 domain-containing transcription repressor RAV2 | ||||
| TrEMBL | A0A2H3ZLG2 | 0.0 | A0A2H3ZLG2_PHODC; AP2/ERF and B3 domain-containing transcription repressor RAV2-like | ||||
| STRING | XP_008782355.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP456 | 38 | 203 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G68840.2 | 1e-116 | related to ABI3/VP1 2 | ||||



