 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PDK_30s1103041g001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Coryphoideae; Phoeniceae; Phoenix
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 292aa MW: 32924.7 Da PI: 5.4284 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 59.6 | 6.8e-19 | 31 | 78 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+WT+eEd++l ++++ +G g+W++ ar+ g++Rt+k+c++rw++yl
PDK_30s1103041g001 31 RGPWTAEEDLILMNYIAAHGEGRWNSLARCAGLKRTGKSCRLRWLNYL 78
89********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 54.2 | 3.3e-17 | 84 | 127 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
rg+ T+eE++l+++++ ++G++ W++Ia++++ gRt++++k++w++
PDK_30s1103041g001 84 RGNITPEEQLLILELHSRWGNR-WSKIAQHLP-GRTDNEIKNYWRT 127
7999******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 17.946 | 26 | 78 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.19E-31 | 28 | 125 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.9E-15 | 30 | 80 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 8.9E-17 | 31 | 78 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 6.2E-24 | 32 | 85 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.33E-11 | 33 | 78 | No hit | No description |
| PROSITE profile | PS51294 | 24.248 | 79 | 133 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.6E-15 | 83 | 131 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.1E-16 | 84 | 127 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.0E-24 | 86 | 132 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 7.62E-12 | 88 | 127 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009620 | Biological Process | response to fungus | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0016036 | Biological Process | cellular response to phosphate starvation | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0001046 | Molecular Function | core promoter sequence-specific DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 292 aa Download sequence Send to blast |
MEMGREVGGE VHGRTYSSAT PPSEEDMDLR RGPWTAEEDL ILMNYIAAHG EGRWNSLARC 60 AGLKRTGKSC RLRWLNYLRP DVRRGNITPE EQLLILELHS RWGNRWSKIA QHLPGRTDNE 120 IKNYWRTRVQ KHAKQLRCDV NSKQFKDVMR YLWMPRLIER IWAASGRSPA TVYQNVSVPM 180 AGDQPAEQGS ESGQVKPSPE TSSAAGSSSD SVGMQFSSPP PVPDCFADCS AGMQEGENKD 240 GDGIHDVQMG GWWPESLPSP GGYGNLGFPD FDQNAWDENL WSVEDIWLQQ QF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 2e-24 | 28 | 131 | 4 | 106 | B-MYB |
| 1h8a_C | 2e-24 | 28 | 131 | 24 | 126 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00334 | DAP | Transfer from AT3G06490 | Download |
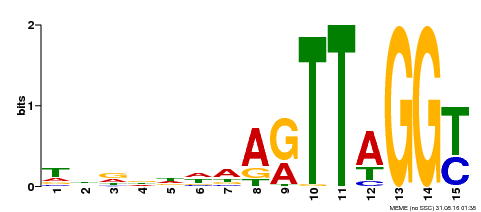
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008781174.1 | 0.0 | transcription factor MYB2 | ||||
| TrEMBL | A0A2H3XBB2 | 0.0 | A0A2H3XBB2_PHODC; transcription factor MYB2 | ||||
| STRING | XP_008781174.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP198 | 38 | 330 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G49620.1 | 3e-85 | myb domain protein 78 | ||||



