 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PDK_30s1103751g030 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Coryphoideae; Phoeniceae; Phoenix
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 624aa MW: 66696.4 Da PI: 8.4141 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 52 | 1.3e-16 | 394 | 440 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K++Ka++L +A++Y+ksLq
PDK_30s1103751g030 394 VHNLSERRRRDRINEKMRALQELIPNC-----NKVDKASMLDEAIDYLKSLQ 440
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 1.5E-20 | 387 | 448 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 9.81E-21 | 388 | 452 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.743 | 390 | 439 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.75E-17 | 393 | 444 | No hit | No description |
| Pfam | PF00010 | 5.4E-14 | 394 | 440 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.2E-18 | 396 | 445 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 624 aa Download sequence Send to blast |
MHGQSNRPRK SSFPTATAAT SSHTGRAQEK ESRDVVTSKI GQIEAVDPLV NDFPPTAASG 60 DAGVNAQDDD DMVPWINYSI EDSLQNDYCS EHMLEFPGMS LNSMSSHTNT AIADRSNGFT 120 QTAKSSRRNV EQGRTSEELA GASDFSRIKS SQLFQASQEW QSLAQSIKPR ATELSTRGTS 180 STHHGLAGCL LSSRPQKQDR AGSRSSQSSS SIDLMNFSHF SRPAALAKAN LQGLDRLRNN 240 EKASTTTSSN PMEPTVIDSI SGLKSILATP GLASAAPEVE QRSSTRRDGN VNNNNSILPG 300 CINRRSSDVA VAAMPSGRNE TEKGPEAVAA SSSVCSGNGA GAASNGPKHT PKRKTYEGEE 360 SGNQREDHED ESMGLRKSAT VRGTSTKRSR AAEVHNLSER RRRDRINEKM RALQELIPNC 420 NKVDKASMLD EAIDYLKSLQ LQVQFLTQIM SMGSGSCMLP LVPPAGIQHM HVPPMAHLSQ 480 MGAGMGMGMR LGYSMGMLDM NGSASCPLLP VPPMHGRQFP FPSIPGTRGL HGMPSSTSLP 540 MFGIPGQGLP ASMPHIPPTC SLSGLPARPN STPGSAGMTN NPSPATDTVP SSTSKDQHQQ 600 SQDLEKIQKP STDDSQMKTS TQVK |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 398 | 403 | ERRRRD |
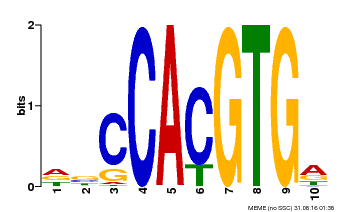
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_026663327.1 | 0.0 | transcription factor PHYTOCHROME INTERACTING FACTOR-LIKE 15-like isoform X1 | ||||
| TrEMBL | A0A3Q0IDA0 | 0.0 | A0A3Q0IDA0_PHODC; transcription factor PHYTOCHROME INTERACTING FACTOR-LIKE 15-like isoform X1 | ||||
| STRING | XP_008800313.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3527 | 37 | 75 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 5e-44 | phytochrome interacting factor 3 | ||||



