 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PDK_30s1145341g001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Coryphoideae; Phoeniceae; Phoenix
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 549aa MW: 59187.5 Da PI: 8.7945 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 16.3 | 2.8e-05 | 57 | 79 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+ C+ C+k F r nL+ H r H
PDK_30s1145341g001 57 FICEVCNKGFQREQNLQLHRRGH 79
89*******************88 PP
| |||||||
| 2 | zf-C2H2 | 13.2 | 0.00026 | 133 | 155 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+kC++C+k++ +s+ k H +t+
PDK_30s1145341g001 133 WKCEKCSKRYAVQSDWKAHSKTC 155
58*****************9998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.30.160.60 | 6.1E-6 | 56 | 79 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 2.95E-7 | 56 | 79 | No hit | No description |
| SMART | SM00355 | 0.0052 | 57 | 79 | IPR015880 | Zinc finger, C2H2-like |
| Pfam | PF12171 | 2.7E-5 | 57 | 79 | IPR022755 | Zinc finger, double-stranded RNA binding |
| PROSITE profile | PS50157 | 10.99 | 57 | 79 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 59 | 79 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 100 | 98 | 128 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 9.1E-5 | 121 | 154 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 2.95E-7 | 128 | 153 | No hit | No description |
| SMART | SM00355 | 130 | 133 | 153 | IPR015880 | Zinc finger, C2H2-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 549 aa Download sequence Send to blast |
MASASSTAPF FEIREEELHN QMKQQQMASI PTSSTAASQT TAPVKKKRNT LMTTNRFICE 60 VCNKGFQREQ NLQLHRRGHN LPWKLRQRST KEVRRRVYVC PEPTCVHHDP SRALGDLTGV 120 KKHFFRKHGE KKWKCEKCSK RYAVQSDWKA HSKTCGTREY RCDCGTLFSR RDSFITHRAF 180 CDALAQEGTR LPTGLSAIGS HFYGNSSMAL GLSQVNSQIT SLQDQSQPSS NIPQLGGSSA 240 SGSHFDHLIS FPNLSPFHPP QPPPSSAFFL GDCSNHEFNE ETQSHHSYPQ NKTFRGLMQL 300 PDLQSSTIAS SSSAAAATAN LFNLSFFSNS SCTSSINTSN NASNQNNHLL ISDQLRNGNG 360 TTLISGNMSN HMGASISSIF STSMQNESSY PQMSATALLQ KAAQMGSTTS GGSSLLQCFN 420 SSSSSGAKRT NFQASFSGGS SGGGDGLRYQ MENETRLHDL MHSLANGKAG LIGGSDGIAA 480 FGGSCTTGQQ GQESGGFGGF NSGLCNVDEA KFHHNLSAGS VRGSDQLTRD FLGVGDMVRS 540 MGGGVSQGE |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3h_C | 6e-36 | 129 | 191 | 3 | 65 | Zinc finger protein JACKDAW |
| 5b3h_F | 6e-36 | 129 | 191 | 3 | 65 | Zinc finger protein JACKDAW |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00255 | DAP | Transfer from AT2G02070 | Download |
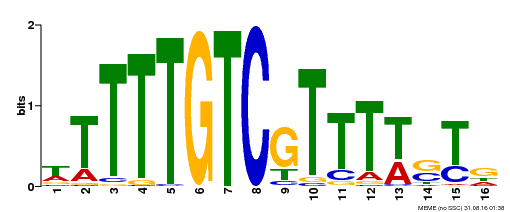
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK374308 | 1e-117 | AK374308.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv3060C18. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017696127.1 | 0.0 | protein indeterminate-domain 5, chloroplastic-like | ||||
| Refseq | XP_017696128.1 | 0.0 | protein indeterminate-domain 5, chloroplastic-like | ||||
| Refseq | XP_026656993.1 | 0.0 | protein indeterminate-domain 5, chloroplastic-like | ||||
| TrEMBL | A0A2H3ZI74 | 0.0 | A0A2H3ZI74_PHODC; protein indeterminate-domain 5, chloroplastic-like | ||||
| STRING | XP_008811073.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1733 | 37 | 98 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G02070.1 | 1e-100 | indeterminate(ID)-domain 5 | ||||



