 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PDK_30s1175051g002 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Coryphoideae; Phoeniceae; Phoenix
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 527aa MW: 56540 Da PI: 6.6338 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 79.2 | 4.4e-25 | 272 | 326 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
Dg++WrKYGqK+ kg+++pr+YYrCt+a gCpv +r+aed +++++tYeg+Hnh+
PDK_30s1175051g002 272 TDGCQWRKYGQKMAKGNPCPRAYYRCTMAtGCPV----QRCAEDRSILITTYEGNHNHP 326
7***************************999*95....67999***************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 4.9E-27 | 256 | 328 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 4.84E-23 | 266 | 328 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 24.237 | 266 | 328 | IPR003657 | WRKY domain |
| SMART | SM00774 | 3.1E-30 | 271 | 327 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 5.3E-21 | 273 | 326 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 527 aa Download sequence Send to blast |
MDSGDHFPPS IEFPVTLNYR RGDTAEEPPG EKRVDEMDFF SDEKKKKRGR DLDFKVPSLS 60 IKKEDLTIDM RFPYTGLNLL TANTGSDQSM VDDGLSPPED NKEGKSKLSS MQAELVRMKE 120 ENQRLKGVLN QVTTNYNALQ MHLVALMQER TQKNGSSQDH EASDEKTDGK IVPRQFLDLG 180 PAANTDEPSR SSIEGGGWDR SSSPTNNVEA GSMEYRLNKT NREDSPDPSS EGWNPNKASK 240 LASPKSAEQS QEATMRKARV SVRAGSEAPM ITDGCQWRKY GQKMAKGNPC PRAYYRCTMA 300 TGCPVQRCAE DRSILITTYE GNHNHPLPPA AMAMASTTSA AASMLLSGSM SSTEGLINSN 360 FLSRTVLPCS SSMATISASA PFPTVTLDLT HTPNPLQFQR LPAQFQVPFP SGTPAFGAPP 420 QPPSLPQVFG QAPNNQSTFS GLQMSPNMDG AQFPRPKPPS AASLPDTVSA ATAAITADPS 480 FKAALVAAIT SIIGGGHQPN SNNNSSSANA TSSNGNGKPA NSNFLAA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ayd_A | 8e-18 | 258 | 330 | 1 | 76 | WRKY transcription factor 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). Modulates phosphate homeostasis and Pi translocation by regulating PHO1 expression (PubMed:25733771). {ECO:0000250, ECO:0000269|PubMed:25733771}. | |||||
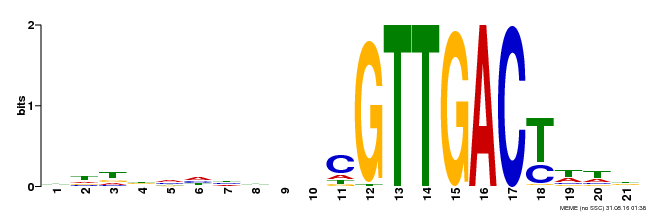
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00428 | DAP | Transfer from AT4G04450 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008779787.1 | 0.0 | probable WRKY transcription factor 31 isoform X1 | ||||
| Swissprot | Q9XEC3 | 1e-133 | WRK42_ARATH; WRKY transcription factor 42 | ||||
| TrEMBL | A0A2H3X801 | 0.0 | A0A2H3X801_PHODC; probable WRKY transcription factor 31 isoform X1 | ||||
| STRING | XP_008779787.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5265 | 36 | 56 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G62300.1 | 1e-117 | WRKY family protein | ||||



