 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PDK_30s697591g002 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Coryphoideae; Phoeniceae; Phoenix
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 239aa MW: 24783.8 Da PI: 10.4983 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 46.6 | 7.3e-15 | 188 | 231 | 5 | 48 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkke 48
+r+rr++kNRe+A rsR+RK+a++ eLe +v++L+++N++L+k+
PDK_30s697591g002 188 RRQRRMIKNRESAARSRARKQAYTMELEAEVAKLKTQNQELQKK 231
79***************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 5.5E-10 | 184 | 236 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 10.909 | 186 | 231 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 1.3E-12 | 188 | 232 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 4.06E-11 | 188 | 233 | No hit | No description |
| CDD | cd14707 | 8.11E-27 | 188 | 234 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 8.8E-15 | 188 | 232 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 191 | 206 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009738 | Biological Process | abscisic acid-activated signaling pathway | ||||
| GO:0010255 | Biological Process | glucose mediated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 239 aa Download sequence Send to blast |
MTLEEFLVRA GVVREDMTLA AAPRPIGNSS NTNNNTNSNV FYGELPNSNN NTGLALGFSQ 60 TSRGNGTVVA NTITNASGAN LAIPATGTRP YAAPLGNTAD LGNPQGLRGG GIVGLGDQGV 120 NNGLMPGMVG MGGGGITVGA VGSPANQISA NGLGKGGDLS SLSPVPYVLS GGFRGRKSSG 180 AVEKVVERRQ RRMIKNRESA ARSRARKQAY TMELEAEVAK LKTQNQELQK KQELEIWGD |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in ABA and stress responses and acts as a positive component of glucose signal transduction. Functions as transcriptional activator in the ABA-inducible expression of rd29B. Binds specifically to the ABA-responsive element (ABRE) of the rd29B gene promoter. {ECO:0000269|PubMed:11005831, ECO:0000269|PubMed:15361142, ECO:0000269|PubMed:16284313, ECO:0000269|PubMed:16463099}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
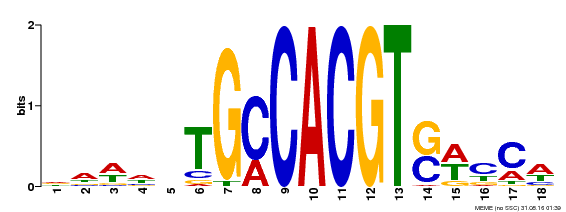
| Motif ID | Method | Source | Motif file |
| MP00186 | DAP | Transfer from AT1G45249 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by drought, salt, abscisic acid (ABA), cold and glucose. {ECO:0000269|PubMed:10636868, ECO:0000269|PubMed:11005831, ECO:0000269|PubMed:15361142, ECO:0000269|PubMed:16284313, ECO:0000269|PubMed:16463099}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008797273.1 | 1e-162 | bZIP transcription factor TRAB1 isoform X1 | ||||
| Refseq | XP_008797274.1 | 1e-162 | bZIP transcription factor TRAB1 isoform X1 | ||||
| Refseq | XP_017699628.1 | 1e-162 | bZIP transcription factor TRAB1 isoform X1 | ||||
| Swissprot | Q9M7Q4 | 2e-52 | AI5L5_ARATH; ABSCISIC ACID-INSENSITIVE 5-like protein 5 | ||||
| TrEMBL | A0A2H3YDQ5 | 1e-160 | A0A2H3YDQ5_PHODC; bZIP transcription factor TRAB1 isoform X1 | ||||
| STRING | XP_008797272.1 | 1e-160 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP706 | 38 | 147 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G45249.1 | 3e-20 | abscisic acid responsive elements-binding factor 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



