 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PDK_30s726031g002 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Coryphoideae; Phoeniceae; Phoenix
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 298aa MW: 32286.4 Da PI: 10.1691 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 41.6 | 2.9e-13 | 103 | 148 | 2 | 47 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
++WT+eE+ ++ + + lG+g+W+ I+r + ++Rt+ q+ s+ qky
PDK_30s726031g002 103 LPWTEEEHRSFLTGLEILGKGDWRGISRSFVTTRTPTQVASHAQKY 148
69*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50158 | 8.664 | 3 | 18 | IPR001878 | Zinc finger, CCHC-type |
| Gene3D | G3DSA:4.10.60.10 | 6.0E-4 | 4 | 23 | IPR001878 | Zinc finger, CCHC-type |
| PROSITE profile | PS51294 | 15.889 | 96 | 153 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.2E-17 | 99 | 154 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 4.7E-17 | 100 | 152 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 3.5E-9 | 101 | 151 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.2E-9 | 103 | 147 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 2.1E-10 | 104 | 148 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.17E-8 | 104 | 149 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 298 aa Download sequence Send to blast |
MGRKCSHCGN NGHNSRTCST PRGVVGSGGL RLFGVQLLLT YPMKKSLSTE CLASSSYLAS 60 SSASPSSSCS SSLVSIDETT KKISNGYLSD GLLGATPERK KGLPWTEEEH RSFLTGLEIL 120 GKGDWRGISR SFVTTRTPTQ VASHAQKYFL RQNSLNEKKR RSSVFDVFKE PSISSELHAP 180 PTLTTYCGAS SQIIAIDLNS SEQETQLPSS LPLMPNSSAC PLPQSPLVER PRAHLSPLNL 240 ELGISSTRPL DQNNSSPNSL FVGTIRVTKV AGRHRTDSPF GVQRFLKTHN SGRKGYTT |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription repressor that binds to 5'-TATCCA-3' elements in gene promoters. Contributes to the sugar-repressed transcription of promoters containing SRS or 5'-TATCCA-3' elements. Transcription repressor involved in a cold stress response pathway that confers cold tolerance. Suppresses the DREB1-dependent signaling pathway under prolonged cold stress. DREB1 responds quickly and transiently while MYBS3 responds slowly to cold stress. They may act sequentially and complementarily for adaptation to short- and long-term cold stress (PubMed:20130099). {ECO:0000269|PubMed:12172034, ECO:0000269|PubMed:20130099}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00565 | DAP | Transfer from AT5G56840 | Download |
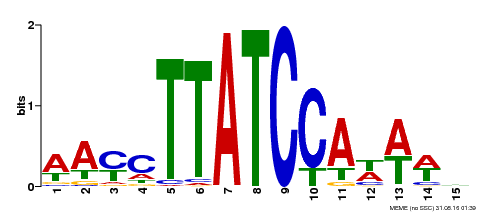
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by sucrose and gibberellic acid (GA) (PubMed:12172034). Induced by cold stress in roots and shoots. Induced by salt stress in shoots. Down-regulated by abscisic aci (ABA) in shoots (PubMed:20130099). {ECO:0000269|PubMed:12172034, ECO:0000269|PubMed:20130099}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008795522.1 | 0.0 | transcription factor KUA1-like | ||||
| Swissprot | Q7XC57 | 1e-47 | MYBS3_ORYSJ; Transcription factor MYBS3 | ||||
| TrEMBL | A0A2H3Y9S8 | 0.0 | A0A2H3Y9S8_PHODC; transcription factor KUA1-like | ||||
| STRING | XP_008795522.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP4231 | 38 | 73 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G56840.1 | 2e-52 | MYB_related family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



