 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PDK_30s752531g002 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Coryphoideae; Phoeniceae; Phoenix
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 297aa MW: 34664.6 Da PI: 8.9189 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 18.6 | 5.3e-06 | 22 | 46 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
++Cp dC+ s++rk++L+rH+ H
PDK_30s752531g002 22 FSCPldDCHLSYRRKDHLTRHLLQH 46
89*******************9887 PP
| |||||||
| 2 | zf-C2H2 | 14.2 | 0.00013 | 51 | 76 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
++Cp +C+++F k n++rH + H
PDK_30s752531g002 51 FTCPveNCNRRFAIKANMNRHVKEfH 76
89********************9988 PP
| |||||||
| 3 | zf-C2H2 | 20.4 | 1.4e-06 | 88 | 112 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
y C+ Cgk+F+ s L++H tH
PDK_30s752531g002 88 YICQepGCGKTFKYASKLRKHEDTH 112
889999***************9998 PP
| |||||||
| 4 | zf-C2H2 | 19.7 | 2.3e-06 | 179 | 203 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirt.H 23
kC+ C ++Fs++snL++Hi+ H
PDK_30s752531g002 179 KCSfkGCQYTFSNRSNLNQHIKAvH 203
688999***************9888 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 3.26E-6 | 17 | 48 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 9.6E-8 | 20 | 48 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.01 | 22 | 46 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 9.328 | 22 | 51 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 24 | 46 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.56E-6 | 49 | 77 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 4.0E-6 | 49 | 77 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0086 | 51 | 76 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 9.868 | 51 | 76 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 53 | 76 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 9.0E-11 | 83 | 113 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 7.42E-7 | 83 | 113 | No hit | No description |
| SMART | SM00355 | 8.7E-4 | 88 | 112 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.344 | 88 | 113 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 90 | 112 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 2.7 | 120 | 145 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 122 | 145 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 8.6 | 148 | 169 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 9.23E-9 | 159 | 216 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 2.8E-8 | 159 | 200 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.946 | 178 | 208 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 4.5E-5 | 178 | 203 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 180 | 203 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 8.7 | 209 | 235 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 211 | 235 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 297 aa Download sequence Send to blast |
MGGPPVKLHQ ESLNHAFPMR PFSCPLDDCH LSYRRKDHLT RHLLQHQGKL FTCPVENCNR 60 RFAIKANMNR HVKEFHEDGC LCEGEKQYIC QEPGCGKTFK YASKLRKHED THAKLDYVEV 120 VCCEPGCMKT FTNTECLKDH IQSCHQYVQC EVCGTQQLKK NLKRHQRMHD GGGVTERIKC 180 SFKGCQYTFS NRSNLNQHIK AVHQELRPFA CRIPGCRNRF PYRHVRDNHE KSGVHVYVQG 240 DFLETDEHLR SRPRGGRKRK CLSVETLQRK RVVPPGQVSS LDDGTYYMRW LLSDDQH |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1tf6_A | 2e-14 | 20 | 205 | 41 | 190 | PROTEIN (TRANSCRIPTION FACTOR IIIA) |
| 1tf6_D | 2e-14 | 20 | 205 | 41 | 190 | PROTEIN (TRANSCRIPTION FACTOR IIIA) |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 251 | 259 | RPRGGRKRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Essential protein (PubMed:22353599). Isoform 1 is a transcription activator the binds both 5S rDNA and 5S rRNA and stimulates the transcription of 5S rRNA gene (PubMed:12711688, PubMed:22353599). Isoform 1 regulates 5S rRNA levels during development (PubMed:22353599). {ECO:0000269|PubMed:12711688, ECO:0000269|PubMed:22353599}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
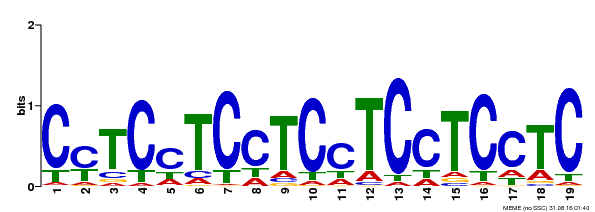
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008793643.1 | 0.0 | transcription factor IIIA-like | ||||
| Swissprot | Q84MZ4 | 1e-80 | TF3A_ARATH; Transcription factor IIIA | ||||
| TrEMBL | A0A2H3Y5C4 | 0.0 | A0A2H3Y5C4_PHODC; transcription factor IIIA-like | ||||
| STRING | XP_008793643.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2382 | 38 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.1 | 8e-84 | transcription factor IIIA | ||||



