 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PDK_30s839411g021 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Coryphoideae; Phoeniceae; Phoenix
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 289aa MW: 32175.1 Da PI: 7.9122 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 94 | 1.1e-29 | 144 | 202 | 1 | 58 |
---SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnh 58
++Dgy+WrKYGqK+ ++++ pr+Y+rC++a +Cpvkkkv+rsaed ++++ tYegeHnh
PDK_30s839411g021 144 VKDGYQWRKYGQKVTRDNPSPRAYFRCSFApSCPVKKKVQRSAEDRSILVATYEGEHNH 202
58***************************99**************************** PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50811 | 26.154 | 139 | 205 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 1.4E-28 | 141 | 205 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 7.32E-25 | 142 | 205 | IPR003657 | WRKY domain |
| SMART | SM00774 | 9.2E-33 | 144 | 204 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 2.6E-24 | 145 | 202 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0002237 | Biological Process | response to molecule of bacterial origin | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0050691 | Biological Process | regulation of defense response to virus by host | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 289 aa Download sequence Send to blast |
MESAWIDHPF LSLDLGVGHL RLPDQAPAED LEAKLNRMSE ENKKLNELLS AIYADYSTLR 60 SQLFDLTSTS PKEKGLVSPS RKRKSESLER ISSDDMTYER TDGVGNRVES ASSEDPCKRF 120 REDPKPKISR VGVRTDPSDS SLVVKDGYQW RKYGQKVTRD NPSPRAYFRC SFAPSCPVKK 180 KVQRSAEDRS ILVATYEGEH NHVHPSQTEV IHGSCQNGSL PCSLSINSSG PTITLDLTQQ 240 GGKDSKEIES PEFRRLLVEQ MASSLTRDPN FTAALATAIS GRILRHPPA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ayd_A | 2e-20 | 143 | 205 | 13 | 74 | WRKY transcription factor 1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 79 | 84 | SRKRKS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription repressor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. Regulates, probably indirectly, the activation of defense-related genes during defense response (By similarity). Modulates plant innate immunity against X.oryzae pv. oryzae (Xoo) (By similarity). {ECO:0000250|UniProtKB:Q6EPZ2, ECO:0000250|UniProtKB:Q6QHD1}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00251 | DAP | Transfer from AT1G80840 | Download |
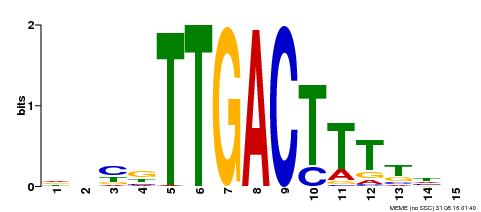
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by biotic elicitors (e.g. fungal chitin oligosaccharide) (By similarity). Induced by pathogen infection (e.g. M.grisea and X.oryzae pv. oryzae (Xoo)) (PubMed:16528562). Accumulates after treatment with benzothiadiazole (BTH) and salicylic acid (SA) (By similarity). {ECO:0000250|UniProtKB:Q6EPZ2, ECO:0000269|PubMed:16528562}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008794348.1 | 0.0 | WRKY transcription factor WRKY76-like | ||||
| Swissprot | Q6IEK5 | 9e-85 | WRK76_ORYSI; WRKY transcription factor WRKY76 | ||||
| TrEMBL | A0A2H3Y6Z1 | 0.0 | A0A2H3Y6Z1_PHODC; WRKY transcription factor WRKY76-like | ||||
| STRING | XP_008794348.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6660 | 35 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G80840.1 | 7e-82 | WRKY DNA-binding protein 40 | ||||



