 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PDK_30s988751g003 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Arecales; Arecaceae; Coryphoideae; Phoeniceae; Phoenix
|
||||||||
| Family | WOX | ||||||||
| Protein Properties | Length: 694aa MW: 78366 Da PI: 7.3038 | ||||||||
| Description | WOX family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 56.1 | 6e-18 | 375 | 436 | 2 | 57 |
T--SS--HHHHHHHHHHHHH..SSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 2 rkRttftkeqleeLeelFek..nrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
++R+ +t++q+++L +l+++ r p+ae+++++++ l +++ ++V++WFqN++a+e++
PDK_30s988751g003 375 STRWIPTADQIRILRDLYYNmgIRSPTAEQIQKISATLrkygKIEGKNVFYWFQNHKARERQ 436
58*****************8779*************************************97 PP
| |||||||
| 2 | Wus_type_Homeobox | 107.2 | 9.9e-35 | 374 | 438 | 2 | 65 |
Wus_type_Homeobox 2 artRWtPtpeQikiLeelyk.sGlrtPnkeeiqritaeLeeyGkiedkNVfyWFQNrkaRerqkq 65
++tRW Pt++Qi+iL++ly+ G+r+P++e+iq+i+a+L++yGkie+kNVfyWFQN+kaRerqk+
PDK_30s988751g003 374 SSTRWIPTADQIRILRDLYYnMGIRSPTAEQIQKISATLRKYGKIEGKNVFYWFQNHKARERQKK 438
789***************9869*****************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF53098 | 5.69E-7 | 3 | 49 | IPR012337 | Ribonuclease H-like domain |
| Pfam | PF07727 | 1.6E-54 | 177 | 330 | IPR013103 | Reverse transcriptase, RNA-dependent DNA polymerase |
| SuperFamily | SSF56672 | 1.16E-8 | 190 | 315 | No hit | No description |
| SuperFamily | SSF46689 | 3.68E-10 | 369 | 441 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 10.074 | 371 | 437 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 7.4E-4 | 373 | 441 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.60E-4 | 376 | 438 | No hit | No description |
| Pfam | PF00046 | 2.4E-15 | 376 | 436 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 3.0E-5 | 377 | 436 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010492 | Biological Process | maintenance of shoot apical meristem identity | ||||
| GO:0080166 | Biological Process | stomium development | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 694 aa Download sequence Send to blast |
MAKSMMHEAG LPKHFWAEAV YTAVYLINRC PTKAVWHQTP FEAWSGRKPS VKHFKVFGCF 60 CYAQIPKEKR YKLDEASEKC IFVGYSSMSK GYRLYSLKTN NIIISRDVIF DEKAKWNWEQ 120 GKVEEQFVPV TISQQSVVQE DQNNDGSSQP LSSSSPPSSP STQSAPSSSS SSPSSTPLKE 180 EVNVEQPQGF IIEGQEEKVY KLKKALYGLK QVPRAWYSKI DSYFNEQGFE KSKSEPTLYV 240 KNQGTTDILI VALYVDDLVL TGSNAEMIEK FKKEMMRKYE MSDLGLLHHF LGIEVYQDEE 300 GVFISQKMYA EKILKKFRMF GCKPIATPLV VNEKLMKEYG RKKYQQQQHE ESNTSSVVGG 360 SVGGSKSNFL CRQSSTRWIP TADQIRILRD LYYNMGIRSP TAEQIQKISA TLRKYGKIEG 420 KNVFYWFQNH KARERQKKRL TADISSTTST PSNSTITTMS PGAVCMVPVL LLGFMVLGRW 480 EAVAVEALCL WKGALELRHT ILFILFVYPY QALTSHINML IYLNLLDGFM VRHPFLWEQE 540 CCMSGGGGDG SGMESSMGWV GLESAATPWM YRYPLEHEAG APMATREIET LPLFPIKQEE 600 EQEGEQEEKK GEFITHCHLN SCDNTTTTSS SNNHNDGGNQ CLPAFYWGGG QAQDQHHLYN 660 HHHLNDGSGG AAAHQAASLD LTLNSYYYAP PGST |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00265 | DAP | Transfer from AT2G17950 | Download |
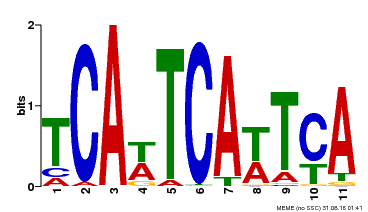
| |||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP12 | 2 | 3 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G17950.1 | 9e-34 | WOX family protein | ||||



