 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PEQU_07312 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; Asparagales; Orchidaceae; Epidendroideae; Vandeae; Aeridinae; Phalaenopsis
|
||||||||
| Family | BBR-BPC | ||||||||
| Protein Properties | Length: 290aa MW: 32175.6 Da PI: 9.8536 | ||||||||
| Description | BBR-BPC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GAGA_bind | 313.2 | 8.9e-96 | 1 | 290 | 1 | 301 |
GAGA_bind 1 mdddgsre.rnkg.yyepaaslkenlglqlmssiaerdakirernlalsekkaavaerd..........maflqrdkalaernkalverdnkllalllv 87
mddd++ +n+g yy+ lk nlglqlm ++ erd+k+ + + e + + +r+ m+f rd + ++ +rd+ l +++
PEQU_07312 1 MDDDATLGiQNWGgYYRTP-PLKGNLGLQLMPTVGERDTKPFFSSV---EAGSGYLHREcgitepstmpMDF-GRDVWYHN------NRDSSKLLHVFQ 88
9999999999***788766.9********************76666...55666777777777778888***.9******9......666554444444 PP
GAGA_bind 88 enslasalpvgvqvlsgtksidslqqlsepqledsavelreeeklealpieeaaeeakekkkkkkrqrakkpkekkakkkkkksekskkkvkkesader 186
n +++ ++ + l+++ s +s q+ l++ ++ +e+ +l ++p++ +++ + kk++k ++++ k +k kk+kk +++k++++k+s +
PEQU_07312 89 GNHH-QHSHSYESLLPEA-SSHSFQM-----LQQVEAPREEKAHLIEEPVVLQPDAEPSKKRSKG--QGRSSKPSKPKKVKKA-KTPKEENAKAS--GK 175
4432.3333455555554.5567888.....4444444334444555555555555555555553..3444455555555552.23444444444..46 PP
GAGA_bind 187 skaekksidlvlngvslDestlPvPvCsCtGalrqCYkWGnGGWqSaCCtttiSvyPLPvstkrrgaRiagrKmSqgafkklLekLaaeGydlsnpvDL 285
+++ kks+d+v+ng++lD s++P+PvC+CtG+++qCY+WG+GGWqSaCCtt+iS yPLP+stkrrgaRiagrKmSqgafkk+LekLa+eG d+s p+DL
PEQU_07312 176 ARSVKKSMDMVINGIDLDLSGMPTPVCTCTGQPQQCYRWGAGGWQSACCTTSISLYPLPMSTKRRGARIAGRKMSQGAFKKVLEKLAGEGEDFSSPIDL 274
89************************************************************************************************* PP
GAGA_bind 286 kdhWAkHGtnkfvtir 301
k +WAkHGtnkfvtir
PEQU_07312 275 KPFWAKHGTNKFVTIR 290
***************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01226 | 7.6E-138 | 1 | 290 | IPR010409 | GAGA-binding transcriptional activator |
| Pfam | PF06217 | 8.2E-101 | 1 | 290 | IPR010409 | GAGA-binding transcriptional activator |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0050793 | Biological Process | regulation of developmental process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 290 aa Download sequence Send to blast |
MDDDATLGIQ NWGGYYRTPP LKGNLGLQLM PTVGERDTKP FFSSVEAGSG YLHRECGITE 60 PSTMPMDFGR DVWYHNNRDS SKLLHVFQGN HHQHSHSYES LLPEASSHSF QMLQQVEAPR 120 EEKAHLIEEP VVLQPDAEPS KKRSKGQGRS SKPSKPKKVK KAKTPKEENA KASGKARSVK 180 KSMDMVINGI DLDLSGMPTP VCTCTGQPQQ CYRWGAGGWQ SACCTTSISL YPLPMSTKRR 240 GARIAGRKMS QGAFKKVLEK LAGEGEDFSS PIDLKPFWAK HGTNKFVTIR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds to GA-rich elements (GAGA-repeats) present in regulatory sequences of genes involved in developmental processes. {ECO:0000250}. | |||||
| UniProt | Transcriptional regulator that specifically binds to GA-rich elements (GAGA-repeats) present in regulatory sequences of genes involved in developmental processes. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00253 | DAP | Transfer from AT2G01930 | Download |
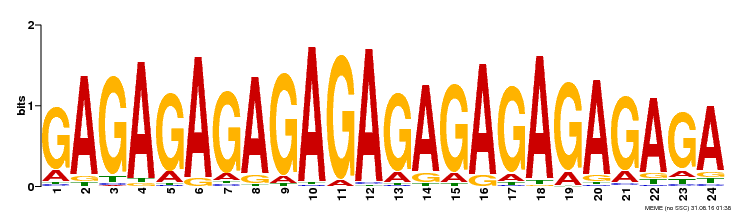
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020575528.1 | 0.0 | protein Barley B recombinant-like | ||||
| Swissprot | P0DH88 | 2e-82 | BBRA_ORYSJ; Barley B recombinant-like protein A | ||||
| Swissprot | P0DH89 | 2e-82 | BBRB_ORYSJ; Barley B recombinant-like protein B | ||||
| TrEMBL | A0A2I0WW46 | 0.0 | A0A2I0WW46_9ASPA; Barley B recombinant-like protein B | ||||
| STRING | XP_008787890.1 | 1e-116 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2211 | 36 | 97 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G01930.2 | 2e-75 | basic pentacysteine1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



