 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PEQU_07664 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; Asparagales; Orchidaceae; Epidendroideae; Vandeae; Aeridinae; Phalaenopsis
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 335aa MW: 35512.2 Da PI: 7.7566 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 74.3 | 1.7e-23 | 217 | 279 | 1 | 63 |
XXXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 1 ekelkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
++elkrerrkq+NRe+ArrsR+RK++e++eL++kv+eL++eN++L+ ele+l k + l++e+
PEQU_07664 217 DRELKRERRKQSNRESARRSRLRKQQECDELARKVAELKGENTSLRMELEQLHKVCRDLEAEN 279
689*******************************************************99987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF07777 | 1.3E-13 | 1 | 86 | IPR012900 | G-box binding protein, multifunctional mosaic region |
| Pfam | PF16596 | 7.6E-8 | 112 | 213 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 3.6E-18 | 214 | 277 | No hit | No description |
| SMART | SM00338 | 1.5E-18 | 217 | 281 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 2.4E-20 | 218 | 279 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 12.714 | 219 | 282 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 4.57E-11 | 220 | 275 | No hit | No description |
| CDD | cd14702 | 1.09E-21 | 222 | 273 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 224 | 239 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010310 | Biological Process | regulation of hydrogen peroxide metabolic process | ||||
| GO:0010629 | Biological Process | negative regulation of gene expression | ||||
| GO:0090342 | Biological Process | regulation of cell aging | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 335 aa Download sequence Send to blast |
MGSPEETSTP AKSPNSTPFS TTATPPPAYN ADWAASVQAF YSGGAAAPTA FFPPVWGGQH 60 LMSPYATPIA YSPMYAPGAV YAHPSMTPGM GYPNVEMGNI KSKGALLKFG EGGKTASGSG 120 EEENSQRSSG GSASEGSSDS GNESAGQQEN SKKRSIGDAF HEGEASNPAI ATPRKRAPKL 180 PVSAPGRTTL PGPTTNLNIG MDIWNPPHAG AAPLKDDREL KRERRKQSNR ESARRSRLRK 240 QQECDELARK VAELKGENTS LRMELEQLHK VCRDLEAENQ HIVEELSSIR GDSSGSLKQG 300 VGDVIDHIPL SSTAISTNTN GKLHHLGGNP DPTSR |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 233 | 239 | RRSRLRK |
| 2 | 233 | 240 | RRSRLRKQ |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00039 | PBM | Transfer from AT4G36730 | Download |
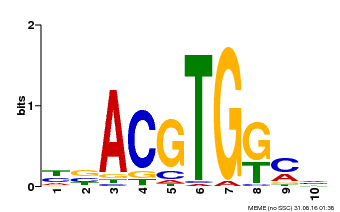
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020581563.1 | 0.0 | G-box-binding factor 1-like isoform X2 | ||||
| TrEMBL | A0A2I0WYD1 | 0.0 | A0A2I0WYD1_9ASPA; G-box-binding factor 1 | ||||
| STRING | XP_008811316.1 | 2e-80 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2160 | 36 | 92 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G36730.1 | 2e-39 | G-box binding factor 1 | ||||



