 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PEQU_09821 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; Asparagales; Orchidaceae; Epidendroideae; Vandeae; Aeridinae; Phalaenopsis
|
||||||||
| Family | GATA | ||||||||
| Protein Properties | Length: 268aa MW: 29374.8 Da PI: 6.1061 | ||||||||
| Description | GATA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GATA | 57.1 | 2.5e-18 | 166 | 200 | 1 | 35 |
GATA 1 CsnCgttkTplWRrgpdgnktLCnaCGlyyrkkgl 35
C +C t kTp+WR gp g+ktLCnaCG+++++ +l
PEQU_09821 166 CLHCETDKTPQWRAGPMGPKTLCNACGVRFKSGRL 200
99*****************************9885 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50114 | 12.229 | 160 | 196 | IPR000679 | Zinc finger, GATA-type |
| SMART | SM00401 | 4.7E-17 | 160 | 210 | IPR000679 | Zinc finger, GATA-type |
| SuperFamily | SSF57716 | 3.52E-15 | 162 | 224 | No hit | No description |
| Gene3D | G3DSA:3.30.50.10 | 1.1E-14 | 163 | 198 | IPR013088 | Zinc finger, NHR/GATA-type |
| CDD | cd00202 | 1.87E-12 | 165 | 207 | No hit | No description |
| Pfam | PF00320 | 3.9E-16 | 166 | 200 | IPR000679 | Zinc finger, GATA-type |
| PROSITE pattern | PS00344 | 0 | 166 | 191 | IPR000679 | Zinc finger, GATA-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009416 | Biological Process | response to light stimulus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 268 aa Download sequence Send to blast |
MMAAEEDGRS EETGGNCSAD SCANSFSGGG SQTRFSDELV YRSLADVGLS GELCEPHEEE 60 AELEWLSNFV EDSFSTEELH KHHLISGLKS ASSPPAHANT CPFTASDDLP PFAVEAPLPG 120 KARSKRSRSA PCDWSSRLRL LSKTATKKNK RPSLAALSPS SDGRRCLHCE TDKTPQWRAG 180 PMGPKTLCNA CGVRFKSGRL VPEYRPALSP TFVLSKHSNS HRKVLELRRQ KEIQEQQQFL 240 LPVSNKGAAE EVEDEFLISG GEVSDEWI |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that specifically binds 5'-GATA-3' or 5'-GAT-3' motifs within gene promoters. May be involved in the regulation of some light-responsive genes (By similarity). Transcription activator involved in xylem formation. Functions upstream of NAC030/VND7, a master switch of xylem vessel differentiation (PubMed:25265867). {ECO:0000250|UniProtKB:Q8LAU9, ECO:0000269|PubMed:25265867}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00530 | DAP | Transfer from AT5G25830 | Download |
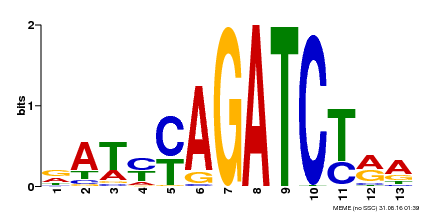
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020575265.1 | 0.0 | GATA transcription factor 9-like | ||||
| Swissprot | P69781 | 1e-62 | GAT12_ARATH; GATA transcription factor 12 | ||||
| TrEMBL | A0A2I0WYE5 | 1e-113 | A0A2I0WYE5_9ASPA; GATA transcription factor 12 | ||||
| STRING | GSMUA_Achr7P25000_001 | 8e-90 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3025 | 31 | 78 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G25830.1 | 2e-57 | GATA transcription factor 12 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



