 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PEQU_09868 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; Asparagales; Orchidaceae; Epidendroideae; Vandeae; Aeridinae; Phalaenopsis
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 291aa MW: 33327.3 Da PI: 7.9237 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 31 | 5.3e-10 | 3 | 37 | 7 | 41 |
HHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 7 errkqkNReAArrsRqRKkaeieeLeekvkeLeae 41
rr+++NReAAr+sR+RKka++++Le+ L++
PEQU_09868 3 IRRLAQNREAARKSRLRKKAYVQQLENSRIRLTQL 37
69************************865555554 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50217 | 8.898 | 1 | 43 | IPR004827 | Basic-leucine zipper domain |
| SMART | SM00338 | 4.1E-4 | 1 | 82 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 4.0E-7 | 3 | 39 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 6.06E-7 | 3 | 43 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 5.2E-8 | 4 | 38 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 4 | 19 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF14144 | 2.6E-32 | 91 | 165 | IPR025422 | Transcription factor TGA like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 291 aa Download sequence Send to blast |
QTIRRLAQNR EAARKSRLRK KAYVQQLENS RIRLTQLEQE LQRARAQGVF FGGGALLGDQ 60 SLPHGINCLS SEATMFDMEF ARWLEEHHRL MCELRAAVQE HLPEHELRLY VDNCLSHYDE 120 LINLKSAITK VDVFHLISGM WKTPAERCFM WMGGFRPSEL IKILLSHIEP LTEQQILGIC 180 GLQQSTQETE EALSQGLEAL HQSLSDTIAS DALSSPSNMA NYMGHMAIAM NKLATLEGFV 240 RQADGLRQQT LHRLHQILTI RQAARCFLSI AEYFHRLRAL SSLWLARPRQ E |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to as1-like elements (5'-TGACGTAAgggaTGACGCA-3') in promoters of target genes. Regulates transcription in response to plant signaling molecules salicylic acid (SA), methyl jasmonate (MJ) and auxin (2,4D) only in leaves. Prevents lateral branching and may repress defense signaling. {ECO:0000269|PubMed:12777042}. | |||||
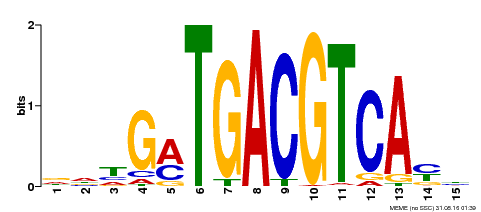
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00491 | DAP | Transfer from AT5G06839 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020579426.1 | 0.0 | bZIP transcription factor TGA10-like isoform X1 | ||||
| Refseq | XP_020579427.1 | 0.0 | bZIP transcription factor TGA10-like isoform X2 | ||||
| Swissprot | Q52MZ2 | 0.0 | TGA10_TOBAC; bZIP transcription factor TGA10 | ||||
| TrEMBL | A0A2I0XFV0 | 0.0 | A0A2I0XFV0_9ASPA; Transcription factor HBP-1b(C38) | ||||
| STRING | XP_010257393.1 | 0.0 | (Nelumbo nucifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP7876 | 35 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G06839.1 | 1e-145 | bZIP family protein | ||||



