 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PEQU_10397 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; Asparagales; Orchidaceae; Epidendroideae; Vandeae; Aeridinae; Phalaenopsis
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 344aa MW: 37818 Da PI: 7.3965 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 30.4 | 8.6e-10 | 180 | 238 | 5 | 63 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
kr++r NR +A rs +RK + eLe+kv++L++e ++L +l l+k + l++e+
PEQU_10397 180 KRAKRILANRQSAARSKERKVRHTNELERKVQTLQTEATTLLAQLTLLQKDSTDLTTEN 238
9***************************************************9999987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 1.1E-15 | 176 | 240 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 10.978 | 178 | 241 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 3.09E-10 | 180 | 231 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 3.4E-9 | 180 | 234 | No hit | No description |
| Pfam | PF00170 | 1.1E-8 | 180 | 238 | IPR004827 | Basic-leucine zipper domain |
| CDD | cd14703 | 2.18E-16 | 181 | 216 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007231 | Biological Process | osmosensory signaling pathway | ||||
| GO:0008272 | Biological Process | sulfate transport | ||||
| GO:0009294 | Biological Process | DNA mediated transformation | ||||
| GO:0009652 | Biological Process | thigmotropism | ||||
| GO:0009970 | Biological Process | cellular response to sulfate starvation | ||||
| GO:0045596 | Biological Process | negative regulation of cell differentiation | ||||
| GO:0051170 | Biological Process | nuclear import | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0003682 | Molecular Function | chromatin binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0043621 | Molecular Function | protein self-association | ||||
| GO:0051019 | Molecular Function | mitogen-activated protein kinase binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 344 aa Download sequence Send to blast |
MDSNRPLETL QKNQRFAGGQ IPPNPRSSGH RRAYSESFRL TEEFLFDSDP DFNFSDIDFP 60 SLSDENKFGS TTTAAAIPIP TDSGGTSKPL AVNPVSRPSV GGAHLRSLSL DTAFFDGLGF 120 QAPGATSGSA QEKKPQHRHS SSMDGSTSPF EGESVPPSSD FSKAAMTADK LAELALIDPK 180 RAKRILANRQ SAARSKERKV RHTNELERKV QTLQTEATTL LAQLTLLQKD STDLTTENRE 240 LKLRLQSMEQ QAHLRDALNE ALSEEVRRLK MATSQVQNID GNTVNSGFQQ RSSPYFCHPQ 300 QFPNWPTQQI PSSSLNSHLS QHLNSTLSPA SPKGTTIHDS MDLM |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds specifically to the VIP1 response elements (VREs) DNA sequence 5'-ACNGCT-3' found in some stress genes (e.g. TRX8 and MYB44), when phosphorylated/activated by MPK3. Required for Agrobacterium VirE2 nuclear import and tumorigenicity. Promotes transient expression of T-DNA in early stages by interacting with VirE2 in complex with the T-DNA and facilitating its translocation to the nucleus, and mediates stable genetic transformation by Agrobacterium by binding H2A histone. Prevents cell differentiation and shoot formation. Limits sulfate utilization efficiency (SUE) and sulfate uptake, especially in low-sulfur conditions. {ECO:0000269|PubMed:11432846, ECO:0000269|PubMed:12124400, ECO:0000269|PubMed:15108305, ECO:0000269|PubMed:15824315, ECO:0000269|PubMed:17947581, ECO:0000269|PubMed:19820165, ECO:0000269|PubMed:20547563}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00184 | DAP | Transfer from AT1G43700 | Download |
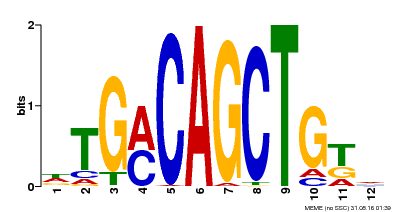
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Transcriptionally activated during the acquisition of pluripotentiality (in protoplasts) by pericentromeric chromatin decondensation and DNA demethylation. Targeted to degradation by the proteasome by VBF and Agrobacterium virF in SCF(VBF) and SCF(virF) E3 ubiquitin ligase complexes after mediating T-DNA translocation to the nucleus. {ECO:0000269|PubMed:15108305}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020573601.1 | 0.0 | transcription factor VIP1-like | ||||
| Swissprot | Q9MA75 | 2e-65 | VIP1_ARATH; Transcription factor VIP1 | ||||
| TrEMBL | A0A2I0W3R6 | 1e-145 | A0A2I0W3R6_9ASPA; Transcription factor VIP1 | ||||
| TrEMBL | A0A2I0WQG8 | 1e-145 | A0A2I0WQG8_9ASPA; Transcription factor VIP1 | ||||
| STRING | XP_008781393.1 | 1e-111 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1925 | 38 | 92 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G43700.1 | 3e-54 | VIRE2-interacting protein 1 | ||||



