 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PEQU_11711 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; Asparagales; Orchidaceae; Epidendroideae; Vandeae; Aeridinae; Phalaenopsis
|
||||||||
| Family | GRAS | ||||||||
| Protein Properties | Length: 572aa MW: 62407.8 Da PI: 5.1413 | ||||||||
| Description | GRAS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GRAS | 450.6 | 1e-137 | 203 | 572 | 1 | 373 |
GRAS 1 lvelLlecAeavssgdlelaqalLarlselaspdgdpmqRlaayfteALaarlarsvselykalppsetseknsseelaalklfsevsPilkfshltaN 99
lv++L++cAeav++++ ++ + l++++s las++g +m+++a yf++ALa+r++r l+p+++ + +s+ + + f+e +P+lkf+h+taN
PEQU_11711 203 LVHALMACAEAVQQENYKASDVLVKQISMLASSQGGAMRKVAGYFAQALARRIYR--------LNPQQDCSLDSAFSDILQMHFYESCPYLKFAHFTAN 293
689****************************************************........88888886666666666667**************** PP
GRAS 100 qaIleavegeervHiiDfdisqGlQWpaLlqaLasRpegppslRiTgvgspesgskeeleetgerLakfAeelgvpfefnvlvakrledleleeLrvkp 198
qaIlea++g++rvH+iDf+++qG+QWpaLlqaLa Rp+gpps+R+Tg+g p+++++++l+++g++La++A++++v+fe++ +va++l+dle+ +L+ p
PEQU_11711 294 QAILEAFAGCRRVHVIDFSMKQGMQWPALLQALALRPGGPPSFRLTGIGLPQPDNTDALQQVGWKLAQLADTIHVDFEYRGFVANSLADLEPYMLESPP 392
***********************************************************************************************7777 PP
GRAS 199 ........gEalaVnlvlqlhrlldesvsleserdevLklvkslsPkvvvvveqeadhnsesFlerflealeyysalfdsleaklpreseerikvErel 289
E++aVn+v++lhrll++++sle+ vL +v ++P++v++veqea+hn+++F+erf+eal++ys++fdslea+ +e ++++++++++
PEQU_11711 393 sanitsdePEVVAVNSVFELHRLLAKPASLEK----VLATVLAVKPRIVTIVEQEANHNTGTFMERFTEALHFYSTMFDSLEAGGFAEGHQDQVMSEVY 487
777777779***********************....*********************************************88877778999999**** PP
GRAS 290 lgreivnvvacegaerrerhetlekWrerleeaGFkpvplsekaakqaklllrkvk.sdgyrveeesgslvlgWkdrpLvsvSaW 373
lgr+i+nvvacegaer+erhetl++Wr r+e aGF+pv+l+++a kqa++ll+ + ++gyrvee++g+l+lgW++rpL+++SaW
PEQU_11711 488 LGRQICNVVACEGAERTERHETLSHWRGRMEGAGFEPVHLGSNAFKQASMLLALFTgGEGYRVEEKEGCLTLGWHTRPLIATSAW 572
************************************************************************************* PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF12041 | 5.4E-31 | 6 | 76 | IPR021914 | Transcriptional factor DELLA, N-terminal |
| SMART | SM01129 | 2.4E-30 | 6 | 82 | No hit | No description |
| PROSITE profile | PS50985 | 67.347 | 177 | 552 | IPR005202 | Transcription factor GRAS |
| Pfam | PF03514 | 3.6E-135 | 203 | 572 | IPR005202 | Transcription factor GRAS |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009867 | Biological Process | jasmonic acid mediated signaling pathway | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010187 | Biological Process | negative regulation of seed germination | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042176 | Biological Process | regulation of protein catabolic process | ||||
| GO:0042538 | Biological Process | hyperosmotic salinity response | ||||
| GO:2000033 | Biological Process | regulation of seed dormancy process | ||||
| GO:2000377 | Biological Process | regulation of reactive oxygen species metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 572 aa Download sequence Send to blast |
DEETNDELLA VLGYKVRSSD MEIVAQKLQQ LEMAMGGSAP HDDELLSHLA VDTVHYNPSD 60 LSNWLENMLS ELSTPPLPLP PVMHPNSVVF DLPPAPILAT MDSTVTSIEL SAPSATMLRS 120 NYALRTTNTS GNRAVYGSET QMDGTGPRDR KRMKTSSSSS SSRGGSGGVI GRAAPSMSST 180 CGSEVSSAMP VVMVDTQEVG IRLVHALMAC AEAVQQENYK ASDVLVKQIS MLASSQGGAM 240 RKVAGYFAQA LARRIYRLNP QQDCSLDSAF SDILQMHFYE SCPYLKFAHF TANQAILEAF 300 AGCRRVHVID FSMKQGMQWP ALLQALALRP GGPPSFRLTG IGLPQPDNTD ALQQVGWKLA 360 QLADTIHVDF EYRGFVANSL ADLEPYMLES PPSANITSDE PEVVAVNSVF ELHRLLAKPA 420 SLEKVLATVL AVKPRIVTIV EQEANHNTGT FMERFTEALH FYSTMFDSLE AGGFAEGHQD 480 QVMSEVYLGR QICNVVACEG AERTERHETL SHWRGRMEGA GFEPVHLGSN AFKQASMLLA 540 LFTGGEGYRV EEKEGCLTLG WHTRPLIATS AW |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3g_A | 3e-62 | 210 | 572 | 26 | 378 | Protein SCARECROW |
| 5b3h_A | 3e-62 | 210 | 572 | 25 | 377 | Protein SCARECROW |
| 5b3h_D | 3e-62 | 210 | 572 | 25 | 377 | Protein SCARECROW |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcriptional regulator that acts as a repressor of the gibberellin (GA) signaling pathway. Probably acts by participating in large multiprotein complexes that repress transcription of GA-inducible genes. Upon GA application, it is degraded by the proteasome, allowing the GA signaling pathway. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00611 | ChIP-seq | Transfer from AT2G01570 | Download |
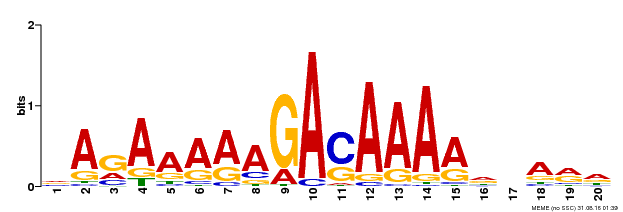
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020591296.1 | 0.0 | LOW QUALITY PROTEIN: DELLA protein DWARF8-like | ||||
| Swissprot | Q9ST48 | 0.0 | DWRF8_MAIZE; DELLA protein DWARF8 | ||||
| TrEMBL | A0A2I0VD31 | 0.0 | A0A2I0VD31_9ASPA; DELLA protein DWARF8 | ||||
| STRING | XP_008782281.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP998 | 38 | 138 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G14920.1 | 0.0 | GRAS family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



