 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PEQU_16338 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; Asparagales; Orchidaceae; Epidendroideae; Vandeae; Aeridinae; Phalaenopsis
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 754aa MW: 81734.9 Da PI: 5.6878 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 68.2 | 1.1e-21 | 60 | 115 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe+lF+++++p++++r eL+k+lgL++rqVk+WFqNrR+++k
PEQU_16338 60 KKRYHRHTPKQIQELEALFKECPHPDEKQRLELSKRLGLETRQVKFWFQNRRTQMK 115
688999***********************************************999 PP
| |||||||
| 2 | START | 171.1 | 7.4e-54 | 269 | 495 | 2 | 206 |
HHHHHHHHHHHHHHHC-TT-EEEE.....EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEEEEEEC CS
START 2 laeeaaqelvkkalaeepgWvkss.....esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdetla....kaetlevis 86
la++a++el+k+a+ +ep+W e +n + + f+++ + + +ea r++++v+ ++ lve+l+d + +W +++ + +t++ is
PEQU_16338 269 LALSAMDELLKMAEMGEPLWIVAGqgngrETLNFKQYEEMFPSGISvkevgFVSEATRETAMVIINSIALVETLMDAA-RWADMFPgiiaRTATTDIIS 366
6899****************98887777766666666666666655888899**************************.******************** PP
TT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--...-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--SSX CS
START 87 sg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppe..sssvvRaellpSgiliepksnghskvtwvehvdlkgrl 176
+g galqlm aelq+lsplvp R++ f+R+++ql + w +vdvSvd +++ + s+ ++++++lpSg+++++++ng+skvtwveh+ ++++
PEQU_16338 367 PGvggsrnGALQLMHAELQVLSPLVPiREVNFLRFCKQLADAAWGVVDVSVDAIRDNHTsaSAASAKCRRLPSGCVVQDMPNGYSKVTWVEHAAYEESA 465
******************************************************9999999999*********************************** PP
XHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 177 phwllrslvksglaegaktwvatlqrqcek 206
h+l+r+l++sgla ga++wvatlqrqce+
PEQU_16338 466 AHQLYRPLLRSGLALGAQRWVATLQRQCEC 495
****************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 5.56E-21 | 47 | 117 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 2.3E-22 | 47 | 111 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 18.091 | 57 | 117 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 2.5E-20 | 58 | 121 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 2.9E-19 | 60 | 115 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 5.30E-20 | 60 | 118 | No hit | No description |
| PROSITE pattern | PS00027 | 0 | 92 | 115 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 41.192 | 259 | 498 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 5.77E-28 | 262 | 495 | No hit | No description |
| CDD | cd08875 | 2.83E-113 | 263 | 494 | No hit | No description |
| SMART | SM00234 | 2.0E-38 | 268 | 495 | IPR002913 | START domain |
| Pfam | PF01852 | 1.4E-45 | 269 | 495 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 3.19E-22 | 523 | 747 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 754 aa Download sequence Send to blast |
QSNLENQGDY NRLASIGGGG GVGELDGICR NKEDENESRS GSDNFEGGSG DDLDENPRKK 60 KRYHRHTPKQ IQELEALFKE CPHPDEKQRL ELSKRLGLET RQVKFWFQNR RTQMKTQMER 120 HENTILRQEN DKLRAESITI RDAMRNPVCS NCGGPAVLGE VSLEEQHLRI ENARLKDELD 180 RVCTLAGKFI GRPISSLASS ISTMPNSSLE LAVGGNAFGA FGPAPHTFSS VTDFVAAASS 240 PLGTVITPPV RASIPPVDRS LERSVFLDLA LSAMDELLKM AEMGEPLWIV AGQGNGRETL 300 NFKQYEEMFP SGISVKEVGF VSEATRETAM VIINSIALVE TLMDAARWAD MFPGIIARTA 360 TTDIISPGVG GSRNGALQLM HAELQVLSPL VPIREVNFLR FCKQLADAAW GVVDVSVDAI 420 RDNHTSASAA SAKCRRLPSG CVVQDMPNGY SKVTWVEHAA YEESAAHQLY RPLLRSGLAL 480 GAQRWVATLQ RQCECLAILM SSSVTNSDHT AIMAAGRRSM LKLAQRMTDN FCAGVCASSA 540 RKWSRLANMG SSIGEDVRVM TRQSMEDPGE PPGVVLSAAT SVWLPVSPNR LFDFLRNESL 600 RSQWDILSNG GPMQEMAHIA KGQDQGNAVS LLRVSAVSAN QSSMLILQET CTDESGSMVV 660 YAPVDVPAMH VVMNGGDSAY VALLPSGFAV MPDGQVAEDG EGKMGGGGSL LTVAFQILVN 720 SLPMAKLTVE SVETVNNLIS CTVQKIKAAL HCEN |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
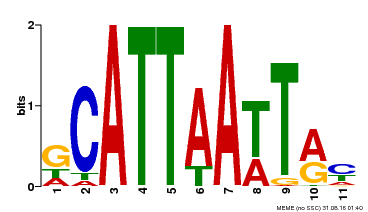
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020585632.1 | 0.0 | homeobox-leucine zipper protein ROC5-like | ||||
| Swissprot | Q6EPF0 | 0.0 | ROC5_ORYSJ; Homeobox-leucine zipper protein ROC5 | ||||
| TrEMBL | A0A2I0X866 | 0.0 | A0A2I0X866_9ASPA; Homeobox-leucine zipper protein ROC5 | ||||
| STRING | XP_008799528.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1203 | 37 | 116 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00730.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



