 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PEQU_19283 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; Asparagales; Orchidaceae; Epidendroideae; Vandeae; Aeridinae; Phalaenopsis
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 582aa MW: 65206.3 Da PI: 5.7906 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 395.4 | 7.6e-121 | 29 | 369 | 2 | 322 |
XXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 2 elkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpvegasdsLraWWkekve 100
l++rmwkd+++lkr++e+ + + +a ++ k +++++qa rkkmsraQDgiLkYMlk +evcna+GfvYgi+p+kgkpv+gasd+LraWWkekv+
PEQU_19283 29 ALSRRMWKDRIKLKRIRENLRL-A--ELQASQKPKAKQPSDQALRKKMSRAQDGILKYMLKLVEVCNARGFVYGIVPDKGKPVSGASDNLRAWWKEKVK 124
589***********88887664.3..4445789999*************************************************************** PP
XXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS-TTS--TTT--HHHH---S--HHHHHHT--TT--.-----GG CS
EIN3 101 fdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdppqrrfplekgvepPWWPtGkelwwgelglskdqgtppykkph 199
fd+ngp ai+ y+ + + +++++ +s++ hsl++lqD+tlgSLLs+lmqhcdppqr++plekg +pPWWP+G+e+ww +lgl+k q pykkph
PEQU_19283 125 FDKNGPSAITVYEEEISATGNKDN----SSKNCHSLMDLQDATLGSLLSSLMQHCDPPQRKYPLEKGHPPPWWPSGNEDWWMNLGLPKGQV-SPYKKPH 218
************766555554444....599***********************************************************9.9****** PP
G--HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT-S--XXXX...XX......XXXXXXXXXXXXXXXXXXXX...X CS
EIN3 200 dlkkawkvsvLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeekecatvsah...ss......slrkqspkvtlsceqkedve...g 286
dlkk wkv+vLt vikhmsp+i+++r + r+sk+lqdkmsakes+++l vl++ee +++++s + s+ sl+ ++++ ++++++dv+ g
PEQU_19283 219 DLKKIWKVGVLTGVIKHMSPNIDKVRTHIRKSKCLQDKMSAKESAIWLGVLSREEMIAQQLSGDnvtSEvtenqqSLQ-EERQDDTNSSSEYDVNdfaG 316
**************************************************************9965322666664444.44455555555777778874 PP
XXXXX.XXXXXXXXXX...................XXXXXXXXXXXXXXXXXXXX CS
EIN3 287 kkeskikhvqavktta...................gfpvvrkrkkkpsesakvss 322
s +++ a kt++ + +vvrk+++ e++ +
PEQU_19283 317 APGSA-SSMGAGKTQHmenqpsleiiastakeqenSSHVVRKKDQ-VNERQRHKR 369
44444.445555554455677777777777777774444444443.222222222 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 5.3E-119 | 29 | 273 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 1.7E-67 | 148 | 280 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 1.96E-58 | 153 | 276 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 582 aa Download sequence Send to blast |
DGLEFEGDDV RCENLTENDV SDEEIDEEAL SRRMWKDRIK LKRIRENLRL AELQASQKPK 60 AKQPSDQALR KKMSRAQDGI LKYMLKLVEV CNARGFVYGI VPDKGKPVSG ASDNLRAWWK 120 EKVKFDKNGP SAITVYEEEI SATGNKDNSS KNCHSLMDLQ DATLGSLLSS LMQHCDPPQR 180 KYPLEKGHPP PWWPSGNEDW WMNLGLPKGQ VSPYKKPHDL KKIWKVGVLT GVIKHMSPNI 240 DKVRTHIRKS KCLQDKMSAK ESAIWLGVLS REEMIAQQLS GDNVTSEVTE NQQSLQEERQ 300 DDTNSSSEYD VNDFAGAPGS ASSMGAGKTQ HMENQPSLEI IASTAKEQEN SSHVVRKKDQ 360 VNERQRHKRP RLDSNPSHQS TGEHQNRDVA LKPENGFSSM NLMDLPLAKD QANGLMQANG 420 LMQANGLISW KSTDGLFATQ TNHVGNQLLF SQPMTNRMFH PSPINVGVQN MIVAGQPLLY 480 TGRQYEEFTA PLVDHPVNPM NVSFPAINPV NRDSLPLNGG SNSAPRDMQP FIEQTYPIDS 540 DKFIGQFDSS VELAMDPFAL SSPLIDIDNF QIDDEALMEY LG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 2e-65 | 156 | 278 | 12 | 134 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00649 | PBM | Transfer from LOC_Os09g31400 | Download |
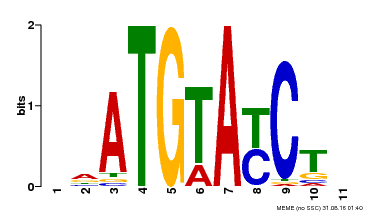
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020582325.1 | 0.0 | LOW QUALITY PROTEIN: ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Swissprot | O23116 | 1e-147 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | A0A2I0VI96 | 0.0 | A0A2I0VI96_9ASPA; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| STRING | XP_008797493.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3720 | 37 | 79 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 1e-146 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



