 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PEQU_19348 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; Asparagales; Orchidaceae; Epidendroideae; Vandeae; Aeridinae; Phalaenopsis
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 269aa MW: 30268.2 Da PI: 8.0553 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 55.9 | 7.1e-18 | 114 | 168 | 2 | 56 |
T--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 2 rkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
rk+ ++tkeq Le+ F+++++++ ++++ LAk+l+L rq+ vWFqNrRa+ k
PEQU_19348 114 RKKLRLTKEQSALLEDRFKEHSTLNPKQKQTLAKQLNLRARQIEVWFQNRRARTK 168
788899***********************************************98 PP
| |||||||
| 2 | HD-ZIP_I/II | 119.5 | 1.8e-38 | 114 | 202 | 1 | 90 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLree 90
+kk+rl+keq++lLE+ F+e+++L+p++K++la++L+l++rq++vWFqnrRARtk+kq+E+d+e+L+r++++l+een+rL ke +eL+ +
PEQU_19348 114 RKKLRLTKEQSALLEDRFKEHSTLNPKQKQTLAKQLNLRARQIEVWFQNRRARTKLKQTEVDCEFLRRCCETLTEENRRLLKELQELK-A 202
69*************************************************************************************9.4 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50071 | 17.265 | 110 | 170 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.1E-14 | 112 | 174 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 1.11E-17 | 113 | 179 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-16 | 114 | 168 | IPR009057 | Homeodomain-like |
| Pfam | PF00046 | 2.8E-15 | 114 | 168 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 2.20E-14 | 115 | 171 | No hit | No description |
| PROSITE pattern | PS00027 | 0 | 145 | 168 | IPR017970 | Homeobox, conserved site |
| Pfam | PF02183 | 7.4E-11 | 170 | 203 | IPR003106 | Leucine zipper, homeobox-associated |
| SMART | SM00340 | 3.9E-21 | 170 | 216 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 269 aa Download sequence Send to blast |
MERGDSCDTD LSLGLGLPTI IRQQQLNPPP PPLEPSLTLS LSEEIYGSTT TPKEARLSSP 60 HSTISSFSTS NIKKEKLDVG GEETEVERGS SSRARERERE REREEEDEAS AAARKKLRLT 120 KEQSALLEDR FKEHSTLNPK QKQTLAKQLN LRARQIEVWF QNRRARTKLK QTEVDCEFLR 180 RCCETLTEEN RRLLKELQEL KAIKFSTPPP PLFMQLPAAT LMVCPSCERV TGAGVGVANK 240 AADGNDISRG TFMTSQQHLF NPFAHSTTC |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 92 | 102 | RARERERERER |
| 2 | 162 | 170 | RRARTKLKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
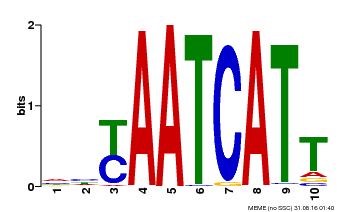
| Motif ID | Method | Source | Motif file |
| MP00477 | DAP | Transfer from AT4G37790 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: In leaves by drought stress. {ECO:0000269|PubMed:17999151}. | |||||
| UniProt | INDUCTION: In leaves by drought stress. {ECO:0000269|PubMed:17999151}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020572230.1 | 0.0 | homeobox-leucine zipper protein HOX19-like | ||||
| Swissprot | A2XE76 | 1e-80 | HOX19_ORYSI; Homeobox-leucine zipper protein HOX19 | ||||
| Swissprot | Q8GRL4 | 1e-80 | HOX19_ORYSJ; Homeobox-leucine zipper protein HOX19 | ||||
| TrEMBL | A0A2I0WRV5 | 1e-146 | A0A2I0WRV5_9ASPA; Homeobox-leucine zipper protein HOX19 | ||||
| STRING | GSMUA_Achr8P26480_001 | 4e-87 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2055 | 36 | 98 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G37790.1 | 3e-63 | HD-ZIP family protein | ||||



