 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PEQU_19589 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; Asparagales; Orchidaceae; Epidendroideae; Vandeae; Aeridinae; Phalaenopsis
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1062aa MW: 121286 Da PI: 7.1203 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 182.2 | 6.1e-57 | 20 | 136 | 3 | 118 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrrcyw 100
+e ++rwl++ ei++iL n++k++++ e+++rp++gsl+L++rk++ryfrkDG++w+kkkdgktv+E+hekLK g+v+vl+cyYah+e+n++fqrr+yw
PEQU_19589 20 REaQHRWLRPAEICEILRNYHKFRIAPEPPNRPSNGSLFLFDRKVLRYFRKDGHNWRKKKDGKTVKEAHEKLKAGSVDVLHCYYAHGEDNENFQRRSYW 118
4449*********************************************************************************************** PP
CG-1 101 lLeeelekivlvhylevk 118
+Leeel +ivlvhy+evk
PEQU_19589 119 MLEEELMHIVLVHYREVK 136
***************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 84.4 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 3.8E-81 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 8.5E-50 | 21 | 134 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 1.4E-7 | 479 | 569 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 2.45E-15 | 481 | 566 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 1.2E-5 | 481 | 566 | IPR002909 | IPT domain |
| CDD | cd00204 | 2.77E-14 | 639 | 770 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 5.1E-18 | 658 | 774 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 17.449 | 669 | 772 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 4.35E-17 | 669 | 772 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 1.1E-7 | 683 | 773 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 0.035 | 711 | 740 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 430 | 750 | 779 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 2.92E-8 | 874 | 931 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.19 | 880 | 902 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.334 | 882 | 910 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 5.5E-4 | 882 | 901 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.001 | 903 | 925 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.597 | 904 | 928 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 1.0E-4 | 906 | 925 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1062 aa Download sequence Send to blast |
MADGRRLGLT PQLDIEQILR EAQHRWLRPA EICEILRNYH KFRIAPEPPN RPSNGSLFLF 60 DRKVLRYFRK DGHNWRKKKD GKTVKEAHEK LKAGSVDVLH CYYAHGEDNE NFQRRSYWML 120 EEELMHIVLV HYREVKGGKA SYLQQREDGE SIRNSNPDSP ACSNSSVNLS QVPSQISDYG 180 SPNSTHQSEY EDADSDNYQS STRYHSVPEV QQYGSWMNVQ LSDTRVPAPT RHDQCDHQRP 240 QAAIPNLDSF TIARDSMTKI ENPSDVGLTF TRSNTKTDIA SWHDMMESCP AGFQTPYDLS 300 VPSSWAATFE DNLERNSFMI QDFLAGDFGT HKEDGTALAS NIIIVWNVVK CPLILILMNG 360 YFVKKQLFLT CFSFFSFFSQ FSANESDSLV ENSQATEENV NQMSDLSNKE HEGLKKYDSF 420 SRWMSKELGE VDDSQINSNS GIYWSSLESE SVVAESNLLN QEQSDTYLMG PLLSQEQLFS 480 IIDFSPNWAY TDLVTKVIVK GKFLMNEEDL QKYKWSCMFG EIEVAANILA NGTLSCYTPP 540 HKSGRVPFYI TCSNRVACSE IREFEYKKDL SRSVGIDDLS NSILQSRFEK LLLLDSMDYS 600 IPACSHLNER SHADCTICSL IMEVDDETIN LFKHDEDFLH VGMKNEDIVS QLKQKLRVWL 660 LHKVGEGDGK GPNVLDEEGQ GVLHLAAALG YDWAIKPTVA AGVNINFRDV HGWTAMHWAA 720 SCGRERTVVT LIALDASPGA LTDPTPEFPT GRTPADLASR NGHKGIAGFL AESSLTNHLR 780 ALNLKGTEKH NLSKPDSLLG FENLPDRINA NLTDGEIQTS LRAVRNATEA AARIFQVFRV 840 QSFNRKQIIK QSDEKFGMSE DRALSLISVK PYKSEQHNMP FHAAAIKIQN KFRGWKGRKE 900 FLITRQRIVK IQAHVRGHQV RKHYKKIIWS VGIVEKVILR WRRKGSGLRG FRPEGLIEGP 960 KDQTIPKKED DDDFLREGRR QTEVRMEKAL ARVKSMVQYP EARDQYRRLV TVVTQLQESK 1020 VYSLFIQFYV WKIYIHTTQF SGIHIMTPKR FTYTPLNSYV FT |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00042 | PBM | Transfer from AT2G22300 | Download |
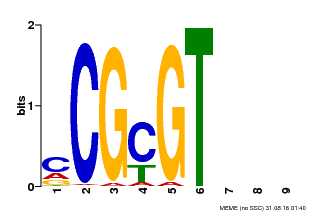
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020588404.1 | 0.0 | calmodulin-binding transcription activator 3-like isoform X2 | ||||
| TrEMBL | A0A2I0VTJ8 | 0.0 | A0A2I0VTJ8_9ASPA; Calmodulin-binding transcription activator 3 | ||||
| STRING | XP_008795547.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP608 | 38 | 140 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22300.2 | 0.0 | signal responsive 1 | ||||



