 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PEQU_26724 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; Asparagales; Orchidaceae; Epidendroideae; Vandeae; Aeridinae; Phalaenopsis
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 324aa MW: 36216 Da PI: 7.9531 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 95.5 | 4.2e-30 | 157 | 212 | 1 | 56 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
k+r++W+peLH++F++a+eqLGGs+ tPk+i+elmkv+ Lt+++vkSHLQkYRl+
PEQU_26724 157 KQRRCWSPELHRKFLDALEQLGGSQSGTPKQIRELMKVERLTNDEVKSHLQKYRLH 212
79****************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 12.814 | 154 | 214 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.7E-28 | 155 | 215 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 5.91E-17 | 155 | 215 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 6.0E-26 | 157 | 212 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 3.2E-7 | 159 | 210 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0055062 | Biological Process | phosphate ion homeostasis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 324 aa Download sequence Send to blast |
MALSARKRDL RAYIEALEDE RRKIQMFSRE LPLCLELITQ AIEIRRQEMA DDEYSGEVPV 60 VEEFISLKPT SSSSPSDKSL KLAGDSKPDW LRSAQLWAPS TVAGVETSPE ESSKLRKPVA 120 KIGGAFQLLE KEKPCPPPAS EVLESSITME NIAAPRKQRR CWSPELHRKF LDALEQLGGS 180 QSGTPKQIRE LMKVERLTND EVKSHLQKYR LHTRRLPPTI QSSSRISPQI PPQFVLVGGI 240 LVPPPEYAVA ASSAEQSSAA FNGLYTRNPQ IKQPKQQKQL NNLPFRPSQS YEDDNNNPAD 300 HESETKSDSC NMSSASQTTT LPLL |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in phosphate homeostasis. Involved in the regulation of the developmental response of lateral roots, acquisition and/or mobilization of phosphate and expression of a subset of genes involved in phosphate sensing and signaling pathway. Is a target of the transcription factor PHR1. {ECO:0000269|PubMed:27016098}. | |||||
| UniProt | Transcriptional repressor that may play a role in response to nitrogen. May be involved in a time-dependent signaling for transcriptional regulation of nitrate-responsive genes. Binds specifically to the DNA sequence motif 5'-GAATC-3' or 5'-GAATATTC-3'. Represses the activity of its own promoter trough binding to these motifs. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00218 | DAP | Transfer from AT1G68670 | Download |
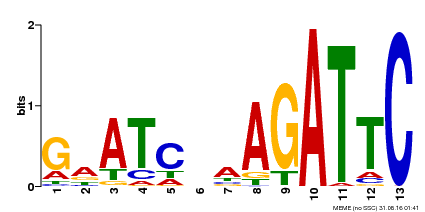
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by nitrate. {ECO:0000269|PubMed:23324170}. | |||||
| UniProt | INDUCTION: Induced under phosphate deprivation conditions. {ECO:0000269|PubMed:27016098}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020585327.1 | 0.0 | myb family transcription factor EFM-like | ||||
| Swissprot | Q6Z869 | 1e-55 | NIGT1_ORYSJ; Transcription factor NIGT1 | ||||
| Swissprot | Q8VZS3 | 5e-56 | HHO2_ARATH; Transcription factor HHO2 | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6470 | 37 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G25550.1 | 4e-51 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



