 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PEQU_36853 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; Asparagales; Orchidaceae; Epidendroideae; Vandeae; Aeridinae; Phalaenopsis
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 306aa MW: 34085.3 Da PI: 9.6297 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 37.5 | 5.5e-12 | 5 | 56 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT.....TTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmg.....kgRtlkqcksrwqky 47
+++WT eE++ l +v ++G g W+tI + +R+ ++k++w++
PEQU_36853 5 KQKWTSEEEDALRAGVDKHGAGKWRTIQKDPEfshclATRSNIDLKDKWRNM 56
79************************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 14.075 | 1 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 9.72E-16 | 2 | 59 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.4E-7 | 4 | 59 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.8E-8 | 5 | 56 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.1E-14 | 5 | 57 | IPR009057 | Homeodomain-like |
| CDD | cd11660 | 1.28E-20 | 6 | 57 | No hit | No description |
| SMART | SM00526 | 4.4E-8 | 109 | 173 | IPR005818 | Linker histone H1/H5, domain H15 |
| PROSITE profile | PS51504 | 18.267 | 112 | 180 | IPR005818 | Linker histone H1/H5, domain H15 |
| Pfam | PF00538 | 3.2E-6 | 115 | 171 | IPR005818 | Linker histone H1/H5, domain H15 |
| SuperFamily | SSF46785 | 2.61E-10 | 115 | 182 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Gene3D | G3DSA:1.10.10.10 | 5.1E-9 | 115 | 174 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006334 | Biological Process | nucleosome assembly | ||||
| GO:0000786 | Cellular Component | nucleosome | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 306 aa Download sequence Send to blast |
MGAPKQKWTS EEEDALRAGV DKHGAGKWRT IQKDPEFSHC LATRSNIDLK DKWRNMIVSA 60 NGQGIREKSR TPKTKALLAP QSSTQSSPIQ APAKFEKQPT AESIKPPQDA KIPSRYNAII 120 MEALTVLQEP NGCDIGAICK YIEQRMEIPS GFRRLLGSKL RRLVAQNKIE KGSRGYQLKD 180 PSFAIKTPTP MQKEPANRLK PLQASGFTKP IDPVEEAATT AAYKIADAEA KAFLASEAVK 240 EAEKISKMAE ETDSLLMLAK EIFERCKLSA CFIILSSRNK ICNSFWDPLT DHTIPSYVFI 300 YLTSEL |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds preferentially double-stranded telomeric repeats, but may also bind to the single telomeric strand. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00652 | PBM | Transfer from LOC_Os01g51154 | Download |
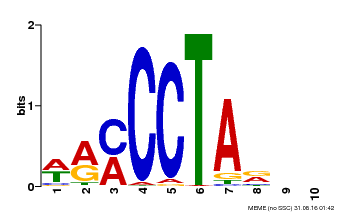
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020596283.1 | 0.0 | single myb histone 4-like | ||||
| Swissprot | Q6WLH4 | 7e-77 | SMH3_MAIZE; Single myb histone 3 | ||||
| TrEMBL | A0A2I0X3P8 | 1e-162 | A0A2I0X3P8_9ASPA; Telomere repeat-binding factor 5 | ||||
| STRING | XP_008797902.1 | 1e-132 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP9247 | 34 | 42 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72740.2 | 6e-51 | MYB_related family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



