 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PGSC0003DMP400006633 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | BBR-BPC | ||||||||
| Protein Properties | Length: 321aa MW: 35925.3 Da PI: 10.3119 | ||||||||
| Description | BBR-BPC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GAGA_bind | 375.9 | 7.1e-115 | 22 | 320 | 18 | 301 |
GAGA_bind 18 aslkenlglqlmssiaerdakirernlalsekkaavaerdmaflqrdkalaernkalverdnkllalllvenslasalpvgvqvlsgtk 106
+s+k q+m+++aerda+i+ernlalsekkaa+aerdma lqrd+a+aern+a++erdn++++l+++en+ +g q+++g+k
PGSC0003DMP400006633 22 PSMK-----QIMAIMAERDAAIQERNLALSEKKAALAERDMAILQRDSAIAERNNAIMERDNAIATLQYRENA-----MTGGQIVRGVK 100
4555.....**************************************************************98.....45788****** PP
GAGA_bind 107 sidslqq.lse.pqledsavelreeeklealpieeaaeeakekkkkkkrqrakkpkekkakkkkkksekskkkvkkesader....... 186
++++ qq +++ p++++++++ re++++ea p++ a+e + +++k rak+pk+ + +kk+ k kkvk+e++d +
PGSC0003DMP400006633 101 HMHHPQQhVHHqPHMGEPTYNPREMHMVEAIPVSPPAPEPAKPRRNK---RAKEPKAVTGSKKTPKA---SKKVKRETEDLNqttygks 183
******9644469*******************999887766666555...55555544443333322...2223333322111345566 PP
GAGA_bind 187 ......................skaekksidlvlngvslDestlPvPvCsCtGalrqCYkWGnGGWqSaCCtttiSvyPLPvstkrrga 253
sk ++k++dl+ln+v++De+t+PvPvCsCtG+lr CYkWGnGGWqS+CCtt++S+yPLP+++++r+a
PGSC0003DMP400006633 184 pewkgaqemvgasddlnrqlavSKPDWKDQDLGLNQVAFDETTMPVPVCSCTGVLRPCYKWGNGGWQSSCCTTNLSMYPLPAVPNKRHA 272
789999*********************************************************************************** PP
GAGA_bind 254 RiagrKmSqgafkklLekLaaeGydlsnpvDLkdhWAkHGtnkfvtir 301
Ri+grKmS++af+klL++LaaeG+dlsnpvDLk+ WAkHGtn+++ti+
PGSC0003DMP400006633 273 RIGGRKMSGSAFTKLLSRLAAEGHDLSNPVDLKNNWAKHGTNRYITIK 320
***********************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01226 | 2.7E-149 | 3 | 320 | IPR010409 | GAGA-binding transcriptional activator |
| Pfam | PF06217 | 5.0E-104 | 10 | 320 | IPR010409 | GAGA-binding transcriptional activator |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 321 aa Download sequence Send to blast |
MSMLRVKSAK FSMYQWLMQH QPSMKQIMAI MAERDAAIQE RNLALSEKKA ALAERDMAIL 60 QRDSAIAERN NAIMERDNAI ATLQYRENAM TGGQIVRGVK HMHHPQQHVH HQPHMGEPTY 120 NPREMHMVEA IPVSPPAPEP AKPRRNKRAK EPKAVTGSKK TPKASKKVKR ETEDLNQTTY 180 GKSPEWKGAQ EMVGASDDLN RQLAVSKPDW KDQDLGLNQV AFDETTMPVP VCSCTGVLRP 240 CYKWGNGGWQ SSCCTTNLSM YPLPAVPNKR HARIGGRKMS GSAFTKLLSR LAAEGHDLSN 300 PVDLKNNWAK HGTNRYITIK * |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds to GA-rich elements (GAGA-repeats) present in regulatory sequences of genes involved in developmental processes. {ECO:0000269|PubMed:14731261}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00540 | DAP | Transfer from AT5G42520 | Download |
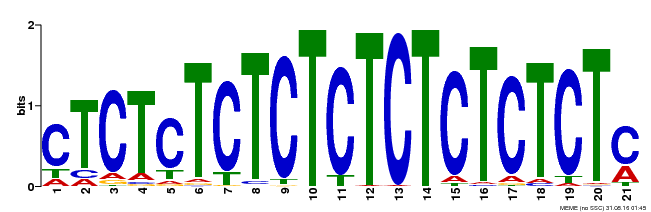
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PGSC0003DMP400006633 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | FJ982314 | 0.0 | FJ982314.1 Solanum tuberosum GAGA-binding transcriptional activator (BBR/BPC3) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001275364.1 | 0.0 | protein BASIC PENTACYSTEINE6-like | ||||
| Refseq | XP_006342016.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6-like isoform X1 | ||||
| Refseq | XP_006342018.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6-like isoform X1 | ||||
| Refseq | XP_015161877.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6-like isoform X1 | ||||
| Swissprot | Q8L999 | 1e-133 | BPC6_ARATH; Protein BASIC PENTACYSTEINE6 | ||||
| TrEMBL | M0ZWF8 | 0.0 | M0ZWF8_SOLTU; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400009534 | 0.0 | (Solanum tuberosum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G42520.1 | 1e-110 | basic pentacysteine 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | PGSC0003DMP400006633 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



