 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PGSC0003DMP400009905 | ||||||||
| Common Name | LOC102604472 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 143aa MW: 15811.1 Da PI: 4.5609 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 25.1 | 4e-08 | 10 | 58 | 3 | 48 |
SS-HHHHHHHHHHHHHTTTT....-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGgg....tWktIartmgkgRtlkqcksrwqkyl 48
W+ eE++ + +a++++ + +W+ a++++ ++t ++k+++q +l
PGSC0003DMP400009905 10 VWSREEEKAFENAIALHWVEdseqQWEQFASMVP-TKTIDELKEHYQLLL 58
6*****************99**************.************875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00717 | 9.6E-4 | 7 | 60 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.9E-6 | 10 | 57 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS50090 | 6.504 | 11 | 58 | IPR017877 | Myb-like domain |
| CDD | cd00167 | 2.29E-6 | 11 | 58 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 3.4E-4 | 11 | 58 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 3.29E-8 | 11 | 64 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 143 aa Download sequence Send to blast |
MSPSDSSSVV WSREEEKAFE NAIALHWVED SEQQWEQFAS MVPTKTIDEL KEHYQLLLED 60 VAAIEAGQVP IPNYKGEEAS SSSTKEVNLG YPGSVDGRRS NCGYSNGFSG TTHDPIGHGG 120 KGNSRSEQER RKGIPWTEEE HR* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2cjj_A | 1e-14 | 1 | 75 | 1 | 74 | RADIALIS |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00191 | DAP | Transfer from AT1G49010 | Download |
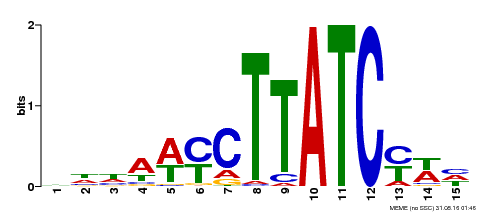
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PGSC0003DMP400009905 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG975442 | 0.0 | HG975442.1 Solanum pennellii chromosome ch03, complete genome. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006341637.1 | 1e-100 | PREDICTED: transcription factor DIVARICATA | ||||
| TrEMBL | M1A420 | 1e-100 | M1A420_SOLTU; Uncharacterized protein | ||||
| TrEMBL | M1A421 | 4e-99 | M1A421_SOLTU; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400014350 | 1e-100 | (Solanum tuberosum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G49010.1 | 2e-32 | MYB family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | PGSC0003DMP400009905 |
| Entrez Gene | 102604472 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



