 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PGSC0003DMP400011136 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 214aa MW: 23565.3 Da PI: 5.1363 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 64.8 | 1.9e-20 | 149 | 198 | 1 | 50 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEE CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfh 50
+Cqv++Cead+se+k yh+rh+vC +++a++v+++g+++r+CqqC+++
PGSC0003DMP400011136 149 RCQVPDCEADISELKGYHKRHRVCLRCANATAVVLDGHSKRYCQQCGKYV 198
6**********************************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 1.1E-18 | 146 | 197 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 17.307 | 147 | 213 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 6.15E-19 | 149 | 201 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 1.1E-14 | 150 | 197 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0055070 | Biological Process | copper ion homeostasis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 214 aa Download sequence Send to blast |
MHNQPSSPSQ TGAPTELRNT QMISSLLSGT PDDPAASSIF DWSDLLEFDL HEQLSISFDD 60 PLHQEQQPET ELVAPVIPSS EDSPQSQDTD AGRIRKRDPR MACSNFLAGQ IPCACPELDE 120 KMEEEEMAGI GPGKKRARTV RASAGAGARC QVPDCEADIS ELKGYHKRHR VCLRCANATA 180 VVLDGHSKRY CQQCGKYVYF FSDEYKKLYA GRV* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul5_A | 2e-26 | 145 | 197 | 1 | 53 | squamosa promoter binding protein-like 7 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that participates in reprogramming global gene expression during copper deficiency in order to improve the metal uptake and prioritize its distribution to copper proteins of major importance (Probable). Binds directly to 5'-GTAC-3' motifs in the microRNA (miRNA) promoter of the stress-responsive miRNAs miR398b and miR398c to activate their transcription. During copper deficiency, activates the copper transporters COPT1 and COPT2, and the copper chaperone CCH, directly or indirectly via miRNAs. Required for the expression of the miRNAs miR397, miR408 and miR857 (PubMed:19122104). Acts coordinately with HY5 to regulate miR408 and its target genes in response to changes in light and copper conditions (PubMed:25516599). Activates miR857 and its target genes in response to low copper conditions (PubMed:26511915). Involved in cadmium stress response by regulating miR397a, miR398b, miR398c and miR857 (PubMed:27352843). Required for iron homeostasis during copper deficiency (PubMed:22374396). {ECO:0000269|PubMed:19122104, ECO:0000269|PubMed:22374396, ECO:0000269|PubMed:25516599, ECO:0000269|PubMed:26511915, ECO:0000269|PubMed:27352843}. | |||||
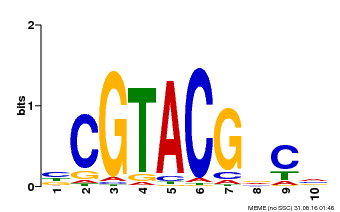
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00604 | PBM | Transfer from AT5G18830 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PGSC0003DMP400011136 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG975440 | 0.0 | HG975440.1 Solanum pennellii chromosome ch01, complete genome. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006356993.1 | 1e-139 | PREDICTED: squamosa promoter-binding-like protein 7 isoform X1 | ||||
| Refseq | XP_006356994.1 | 1e-139 | PREDICTED: squamosa promoter-binding-like protein 7 isoform X2 | ||||
| Swissprot | Q8S9G8 | 2e-49 | SPL7_ARATH; Squamosa promoter-binding-like protein 7 | ||||
| TrEMBL | M1A716 | 1e-158 | M1A716_SOLTU; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400016072 | 1e-139 | (Solanum tuberosum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G18830.2 | 5e-46 | squamosa promoter binding protein-like 7 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | PGSC0003DMP400011136 |



