 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PGSC0003DMP400011976 | ||||||||
| Common Name | LOC102599464 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 272aa MW: 30747.8 Da PI: 6.0061 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 88.4 | 3.9e-28 | 9 | 59 | 1 | 51 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
k ien nrqvtfskRr+g+lKKA ELSvLCd evaviifsstgkl+e+ss
PGSC0003DMP400011976 9 KLIENVNNRQVTFSKRRAGLLKKAGELSVLCDSEVAVIIFSSTGKLFEFSS 59
569**********************************************96 PP
| |||||||
| 2 | K-box | 58 | 4e-20 | 92 | 186 | 7 | 100 |
K-box 7 ks.leeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenkaL 94
+s ++++ ++ q+e++ Lk+e+ +L+ +q +llG+dL+ + l+eL+ Le+qL+++l i+++K+ell++q+e+++k+e++ e +aL
PGSC0003DMP400011976 92 QSqQQTHVLKQEQKEVDSLKDELAKLKMKQLRLLGKDLNGMGLNELRLLEHQLNEGLLAIKERKEELLIQQLEYSRKQEERSALECEAL 180
333555567889***************************************************************************** PP
K-box 95 rkklee 100
r+++ee
PGSC0003DMP400011976 181 RRQVEE 186
***987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50066 | 31.413 | 1 | 61 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 1.2E-39 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| SuperFamily | SSF55455 | 3.27E-31 | 2 | 74 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 2.72E-39 | 2 | 75 | No hit | No description |
| PROSITE pattern | PS00350 | 0 | 3 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 8.1E-28 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 8.2E-25 | 11 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 8.1E-28 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 8.1E-28 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 1.9E-14 | 95 | 184 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 12.266 | 100 | 190 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010047 | Biological Process | fruit dehiscence | ||||
| GO:0010262 | Biological Process | somatic embryogenesis | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048577 | Biological Process | negative regulation of short-day photoperiodism, flowering | ||||
| GO:0060862 | Biological Process | negative regulation of floral organ abscission | ||||
| GO:0060867 | Biological Process | fruit abscission | ||||
| GO:0071365 | Biological Process | cellular response to auxin stimulus | ||||
| GO:2000692 | Biological Process | negative regulation of seed maturation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 272 aa Download sequence Send to blast |
MGRGKIDIKL IENVNNRQVT FSKRRAGLLK KAGELSVLCD SEVAVIIFSS TGKLFEFSST 60 SMKQTLSRYN RCVASTDNSA VEKKSEDNEQ PQSQQQTHVL KQEQKEVDSL KDELAKLKMK 120 QLRLLGKDLN GMGLNELRLL EHQLNEGLLA IKERKEELLI QQLEYSRKQE ERSALECEAL 180 RRQVEELRGL FPLSASLPPP FLEYDRPLEK KYSILKESKE SLDSDTACED GVDDEDSNTT 240 LQLGLPTICR KRKRTEQESP SSNSENQGGS K* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5f28_A | 1e-21 | 1 | 69 | 1 | 69 | MEF2C |
| 5f28_B | 1e-21 | 1 | 69 | 1 | 69 | MEF2C |
| 5f28_C | 1e-21 | 1 | 69 | 1 | 69 | MEF2C |
| 5f28_D | 1e-21 | 1 | 69 | 1 | 69 | MEF2C |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in the negative regulation of flowering, probably through the photoperiodic pathway. Acts as both an activator and a repressor of transcription. Binds DNA in a sequence-specific manner in large CArG motif 5'-CC (A/T)8 GG-3'. Participates probably in the regulation of programs active during the early stages of embryo development. Prevents premature perianth senescence and abscission, fruits development and seed desiccation. Stimulates the expression of at least DTA4, LEC2, FUS3, ABI3, AT4G38680/CSP2 and GRP2B/CSP4. Can enhance somatic embryo development in vitro. {ECO:0000269|PubMed:10318690, ECO:0000269|PubMed:10662856, ECO:0000269|PubMed:12226488, ECO:0000269|PubMed:12743119, ECO:0000269|PubMed:14615187, ECO:0000269|PubMed:15084721, ECO:0000269|PubMed:15686521, ECO:0000269|PubMed:17521410, ECO:0000269|PubMed:18305206, ECO:0000269|PubMed:19269998, ECO:0000269|PubMed:19767455, ECO:0000269|PubMed:8953767}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00508 | DAP | Transfer from AT5G13790 | Download |
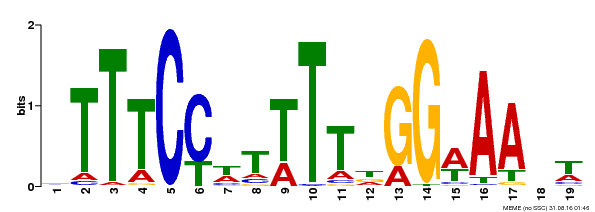
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PGSC0003DMP400011976 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By auxin (2,4-D). Feedback loop leading to direct down-regulation by itself. {ECO:0000269|PubMed:15686521}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT014220 | 0.0 | BT014220.1 Lycopersicon esculentum clone 133400R, mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006345394.1 | 0.0 | PREDICTED: agamous-like MADS-box protein AGL15 | ||||
| Swissprot | Q38847 | 5e-69 | AGL15_ARATH; Agamous-like MADS-box protein AGL15 | ||||
| TrEMBL | M1A8X4 | 0.0 | M1A8X4_SOLTU; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400017416 | 0.0 | (Solanum tuberosum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G13790.1 | 1e-65 | AGAMOUS-like 15 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | PGSC0003DMP400011976 |
| Entrez Gene | 102599464 |



