 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PGSC0003DMP400012934 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 297aa MW: 34771.1 Da PI: 4.9925 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 44.1 | 4.9e-14 | 60 | 105 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+ T +E+ +++++ + lG++ W++I ++++ gRt++++k++w+++l
PGSC0003DMP400012934 60 RGNITSDEEAIIIKLRATLGNR-WSLIVEHLP-GRTDNEIKNYWNSHL 105
7999******************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50090 | 4.368 | 33 | 54 | IPR017877 | Myb-like domain |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-9 | 34 | 67 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.0E-18 | 41 | 114 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 22.159 | 55 | 109 | IPR017930 | Myb domain |
| SMART | SM00717 | 2.0E-14 | 59 | 107 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.6E-12 | 60 | 105 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.27E-8 | 64 | 105 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 5.9E-19 | 68 | 105 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 297 aa Download sequence Send to blast |
MKVDNKRNIK MTKEIKKKKK KIKEEHKMTE MIKEIQQRLL RCGKSCRLRW INYLKTDLKR 60 GNITSDEEAI IIKLRATLGN RWSLIVEHLP GRTDNEIKNY WNSHLSRKVD SLRIPSDEKL 120 PQAVVELAKK GTQQHISKRG RRRKINTSVS NSTNEITVNN DIEFDRTDDK MLWDDDIMLM 180 DDKYGELDQD FITNCLWDGE GENLEIANDD NNNNNNNNEI LSEDNYGKLD CWDWEYLGEV 240 LNNEIWTNST KEAEQENNNN MSLCDINDNM QNCMNKEATP ENIDPMQSDL VTWLLS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 7e-18 | 24 | 111 | 14 | 110 | B-MYB |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00627 | PBM | Transfer from PK06182.1 | Download |
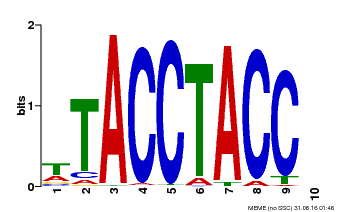
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PGSC0003DMP400012934 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006355861.1 | 0.0 | PREDICTED: transcription repressor MYB6-like | ||||
| TrEMBL | M1AB80 | 0.0 | M1AB80_SOLTU; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400018841 | 0.0 | (Solanum tuberosum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G49330.1 | 4e-38 | myb domain protein 111 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | PGSC0003DMP400012934 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



