 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PGSC0003DMP400019518 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 287aa MW: 31704 Da PI: 9.4228 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 44.7 | 3e-14 | 25 | 69 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r +WT+eE++++++a +++ + Wk+I +g +t q++s+ qky
PGSC0003DMP400019518 25 RESWTEEEHDKFLEALQLFDRD-WKKIEDFVG-SKTVIQIRSHAQKY 69
789*****************77.*********.*************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.12E-17 | 19 | 75 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 16.291 | 20 | 74 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 8.3E-19 | 23 | 72 | IPR006447 | Myb domain, plants |
| Gene3D | G3DSA:1.10.10.60 | 2.6E-7 | 23 | 75 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.1E-11 | 24 | 72 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 6.7E-12 | 25 | 69 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.04E-9 | 27 | 70 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0032922 | Biological Process | circadian regulation of gene expression | ||||
| GO:0043966 | Biological Process | histone H3 acetylation | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0048573 | Biological Process | photoperiodism, flowering | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 287 aa Download sequence Send to blast |
MANNSTPTDA SSKKIRKPYT ITKSRESWTE EEHDKFLEAL QLFDRDWKKI EDFVGSKTVI 60 QIRSHAQKYF LKVQKNGSIA HVPPPRPKRK AAHPYPQKAP KNVLVPLQAS MGYPSSMNSF 120 PPGYPLWDDA SVLINSPSGG VMPSQDEYHL QRIQADIGSK GATLISNSSM SGIRSSNRTA 180 LSSELPDQSK LGSVPHGIPD FAEVYSFIGS VFDPDTRGHV QKLKEMDPIN FETVLMLMRN 240 LTMNLSNPDF EPIKNVLSTY DLSSKVVGLP TGGAVNNRND LSCQTI* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator of evening element (EE)-containing clock-controlled genes. Forms a negative feedback loop with APRR5. Regulates the pattern of histone H3 acetylation of the TOC1 promoter. {ECO:0000269|PubMed:21205033, ECO:0000269|PubMed:21474993, ECO:0000269|PubMed:21483796, ECO:0000269|PubMed:23638299}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00338 | DAP | Transfer from AT3G09600 | Download |
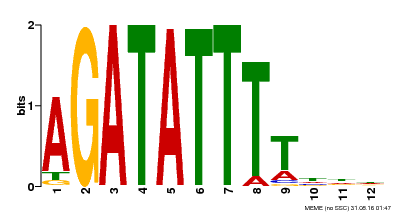
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PGSC0003DMP400019518 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Circadian-regulation. Peak of expression in the afternoon. Down-regulated by cold. {ECO:0000269|PubMed:21205033, ECO:0000269|PubMed:22902701, ECO:0000269|PubMed:23638299}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006339055.1 | 0.0 | PREDICTED: protein REVEILLE 8-like | ||||
| Swissprot | Q8RWU3 | 1e-126 | RVE8_ARATH; Protein REVEILLE 8 | ||||
| TrEMBL | M1ARJ8 | 0.0 | M1ARJ8_SOLTU; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400028711 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA998 | 24 | 78 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G09600.1 | 1e-107 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | PGSC0003DMP400019518 |



