 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PGSC0003DMP400020363 | ||||||||
| Common Name | LOC102589616 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 987aa MW: 111180 Da PI: 5.0342 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 43.1 | 9.9e-14 | 8 | 53 | 2 | 48 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
g W Ede+l+ av ++G+++W++I++ + ++++kqck rw+ +l
PGSC0003DMP400020363 8 GVWKNTEDEILKAAVMKYGKNQWARISSLLV-RKSAKQCKARWYEWL 53
78*****************************.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 32.8 | 1.6e-10 | 60 | 103 | 2 | 48 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
WT eEde+l+++ k++++ W+tIa +g Rt+ qc +r+ k+l
PGSC0003DMP400020363 60 TEWTREEDEKLLHLAKLMPTQ-WRTIAPIVG--RTPSQCLERYEKLL 103
68*****************99.********8..**********9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 22.008 | 2 | 57 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.7E-18 | 5 | 56 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 7.0E-15 | 6 | 55 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.98E-12 | 10 | 53 | No hit | No description |
| Pfam | PF13921 | 4.7E-13 | 10 | 70 | No hit | No description |
| SuperFamily | SSF46689 | 5.39E-21 | 33 | 109 | IPR009057 | Homeodomain-like |
| CDD | cd11659 | 1.97E-30 | 55 | 106 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 3.4E-14 | 57 | 104 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 16.443 | 58 | 107 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.7E-12 | 58 | 105 | IPR001005 | SANT/Myb domain |
| Pfam | PF11831 | 6.6E-58 | 406 | 648 | IPR021786 | Pre-mRNA splicing factor component Cdc5p/Cef1 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009870 | Biological Process | defense response signaling pathway, resistance gene-dependent | ||||
| GO:0010204 | Biological Process | defense response signaling pathway, resistance gene-independent | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0009507 | Cellular Component | chloroplast | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 987 aa Download sequence Send to blast |
MRIMIKGGVW KNTEDEILKA AVMKYGKNQW ARISSLLVRK SAKQCKARWY EWLDPSIKKT 60 EWTREEDEKL LHLAKLMPTQ WRTIAPIVGR TPSQCLERYE KLLDAACTKD ENYDPNDDPR 120 KLKPGEIDPN PESKPARPDP VDMDEDEKEM LSEARARLAN TRGKKAKRKA REKQLEEARR 180 LASLQKRREL KAAGIDVRQR KRKRRGIDYN AEIPFEKKPP PGFYDITEED RPVDQPKFPT 240 TIEELEGERR VDKEARLRKQ DVARNKIAER QDAPTSILHA NKLNDPEAVR KRSKLNLPAP 300 QIPDHELEAI AKIGIASDLI GGDELSEGNA ATRALLANYA QTPQHAMTPM RTPQRTPSTK 360 QDSIMMEAEN QRRLTQSQTP LLGGDNPLLH PSDFSGVTPK KREVQTPNPL LTPSATPGAT 420 SLTPRIGMTP SRDSYGMTPK GTPMRDELHI NEEMDMHNNA KLGQFNSKKE LLSGLKSLPQ 480 PKNEYQIVVQ QPPEENEEPE EKIEEDMSDR IAREKAEEEA KRQALLRKRS KVLQRELPRP 540 PIASLELIKS SLMRADEDKS SFVPPTLIEQ ADEMIRKELV SLLEHDNTKY PLDEKPEKEK 600 KKGVKRKIVA EPAIEDFEED ELKEADGLIK DEAHFLRVAM GHESESLDEF VEVHKTTLND 660 IMYFPTRNAY GLSSVAGNME KLAALQNEFE NVKKKMDDDT KKATKLEQKI KVLTNGYQIR 720 AGKLWSQIES TFKKMDTAGT ELECFRALQK QEQLAASHRI NNMWEEVQKQ KELERTLQKR 780 YGDLIADTQK IQHLMDEYRI QDQMQEEIAA KNRALELAKA EIAEKESIPS ADDVEPSGTV 840 QCSNTEENSA SASHVPIEAD VHAEPSGTDQ CSNAEENSAS IEADNVHVEP SGTSQCPIAE 900 ETSASVSHDT TPQDVEGQVQ VADVSTMDAE ALSDHVPMEG QQNLVEESNT DVTKTEDSTV 960 AAGDVDTKTD DSVVVAGDGE ADPKNM* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5mqf_L | 0.0 | 2 | 792 | 3 | 799 | Cell division cycle 5-like protein |
| 5xjc_L | 0.0 | 2 | 792 | 3 | 799 | Cell division cycle 5-like protein |
| 5yzg_L | 0.0 | 2 | 792 | 3 | 799 | Cell division cycle 5-like protein |
| 5z56_L | 0.0 | 2 | 792 | 3 | 799 | Cell division cycle 5-like protein |
| 5z57_L | 0.0 | 2 | 792 | 3 | 799 | Cell division cycle 5-like protein |
| 5z58_L | 0.0 | 2 | 792 | 3 | 799 | Cell division cycle 5-like protein |
| 6ff4_L | 0.0 | 2 | 792 | 3 | 799 | Cell division cycle 5-like protein |
| 6ff7_L | 0.0 | 2 | 792 | 3 | 799 | Cell division cycle 5-like protein |
| 6icz_L | 0.0 | 2 | 792 | 3 | 799 | Cell division cycle 5-like protein |
| 6id0_L | 0.0 | 2 | 792 | 3 | 799 | Cell division cycle 5-like protein |
| 6id1_L | 0.0 | 2 | 792 | 3 | 799 | Cell division cycle 5-like protein |
| 6qdv_O | 0.0 | 2 | 792 | 3 | 799 | Cell division cycle 5-like protein |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 185 | 204 | KRRELKAAGIDVRQRKRKRR |
| 2 | 198 | 204 | QRKRKRR |
| 3 | 199 | 204 | RKRKRR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
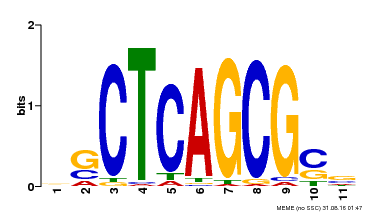
| UniProt | Component of the MAC complex that probably regulates defense responses through transcriptional control and thereby is essential for plant innate immunity. Possesses a sequence specific DNA sequence 'CTCAGCG' binding activity. Involved in mRNA splicing and cell cycle control. May also play a role in the response to DNA damage. {ECO:0000250|UniProtKB:Q99459, ECO:0000269|PubMed:17298883, ECO:0000269|PubMed:17575050, ECO:0000269|PubMed:8917598}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00028 | SELEX | Transfer from AT1G09770 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PGSC0003DMP400020363 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU095496 | 0.0 | EU095496.1 Solanum lycopersicum CDC5-like protein (cdc5) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006361426.1 | 0.0 | PREDICTED: cell division cycle 5-like protein | ||||
| Swissprot | P92948 | 0.0 | CDC5L_ARATH; Cell division cycle 5-like protein | ||||
| TrEMBL | M1ATK6 | 0.0 | M1ATK6_SOLTU; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400029953 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA6139 | 24 | 31 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09770.1 | 0.0 | cell division cycle 5 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | PGSC0003DMP400020363 |
| Entrez Gene | 102589616 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



