 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PGSC0003DMP400021690 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | Nin-like | ||||||||
| Protein Properties | Length: 689aa MW: 75529.9 Da PI: 6.1824 | ||||||||
| Description | Nin-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | RWP-RK | 93.1 | 2e-29 | 288 | 339 | 1 | 52 |
RWP-RK 1 aekeisledlskyFslpikdAAkeLgvclTvLKriCRqyGIkRWPhRkiksl 52
aek+isle+l++yF++++kdAAk+Lgvc+T++KriCRq+GI+RWP+Rki+++
PGSC0003DMP400021690 288 AEKTISLEVLQQYFAGSLKDAAKSLGVCPTTMKRICRQHGISRWPSRKINKV 339
589***********************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51519 | 17.655 | 276 | 359 | IPR003035 | RWP-RK domain |
| Pfam | PF02042 | 2.1E-26 | 291 | 339 | IPR003035 | RWP-RK domain |
| SuperFamily | SSF54277 | 6.87E-24 | 592 | 679 | No hit | No description |
| SMART | SM00666 | 1.3E-24 | 593 | 675 | IPR000270 | PB1 domain |
| Gene3D | G3DSA:3.10.20.240 | 9.7E-27 | 593 | 674 | No hit | No description |
| PROSITE profile | PS51745 | 25.665 | 593 | 675 | IPR000270 | PB1 domain |
| CDD | cd06407 | 2.89E-39 | 594 | 674 | No hit | No description |
| Pfam | PF00564 | 2.6E-19 | 594 | 674 | IPR000270 | PB1 domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0010118 | Biological Process | stomatal movement | ||||
| GO:0010167 | Biological Process | response to nitrate | ||||
| GO:0042128 | Biological Process | nitrate assimilation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 689 aa Download sequence Send to blast |
MNQICNEGRQ NALVEILEIL TAVCETYKLP LAQTWVPCRH RSVLADGGGF KKSCSSFDGS 60 CMGQVCMSTT DVAFYVVDAH MWGFREACAE HHLQKGQGVA GRAYASQKSC FCEDIGQFCK 120 TEYPLVHYAR LFGLSSCLAI CLRSTHTGND DYILEFFLPP NDGDYTDQLA LLNSLLLTMK 180 QHFRSLRVAS GEELEHNWGS VEIIKASTEE KLGSRFDSVP TTKSLPQSAS VANGRTHPDL 240 MEEQQSPVAL NVAKGAEGVN GTAEAHNHAS VPENKQTGKK SERKRGKAEK TISLEVLQQY 300 FAGSLKDAAK SLGVCPTTMK RICRQHGISR WPSRKINKVN RSLSKLKRVI ESVQGADGTF 360 SLTSLAPNSL PVAVGSISWP AGINGSPCKA SEYQEEKNEF SNHGTPGSHE EAEPMDQMLG 420 SRIIGNEELS PKQNGFVREG SHRSRTGSFS REESTGTPTS HGSCQGSPSP ANESSPQNEL 480 VNSPTQESVM KVEGSLEPAR QTTGEINLST SFLMPGLYIP EHTQQQFRGM LVEDAGSSHD 540 LRNLCPAGEA MFDERVPEYS WTNPPCSNGI ATNQVPLPVE KMPQFSSRPE VTSVTIKATY 600 REDIIRFRLC LNSGIYKLKE EVAKRLKLEM GTFDIKYLDD DHEWVLITCD ADLQECIDIS 660 RSSGSNVVRL LVHDIMPNLG SSCESSGE* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in regulation of nitrate assimilation and in transduction of the nitrate signal. {ECO:0000269|PubMed:18826430}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00449 | DAP | Transfer from AT4G24020 | Download |
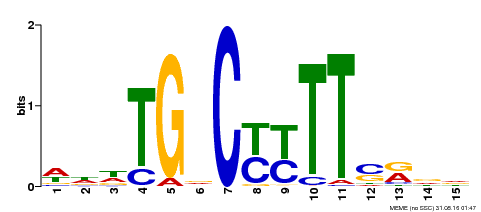
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PGSC0003DMP400021690 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Not regulated by the N source or by nitrate. {ECO:0000269|PubMed:18826430}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG975447 | 0.0 | HG975447.1 Solanum pennellii chromosome ch08, complete genome. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006343817.1 | 0.0 | PREDICTED: protein NLP6-like | ||||
| Swissprot | Q84TH9 | 0.0 | NLP7_ARATH; Protein NLP7 | ||||
| TrEMBL | M1AWJ8 | 0.0 | M1AWJ8_SOLTU; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400031945 | 0.0 | (Solanum tuberosum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G24020.1 | 0.0 | NIN like protein 7 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | PGSC0003DMP400021690 |



