 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PGSC0003DMP400021691 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | Nin-like | ||||||||
| Protein Properties | Length: 640aa MW: 70504.9 Da PI: 6.2713 | ||||||||
| Description | Nin-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | RWP-RK | 23.6 | 1e-07 | 607 | 632 | 1 | 26 |
RWP-RK 1 aekeisledlskyFslpikdAAkeLg 26
aek+isle+l++yF++++kdAAk+Lg
PGSC0003DMP400021691 607 AEKTISLEVLQQYFAGSLKDAAKSLG 632
589*********************98 PP
| |||||||
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0010118 | Biological Process | stomatal movement | ||||
| GO:0010167 | Biological Process | response to nitrate | ||||
| GO:0042128 | Biological Process | nitrate assimilation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 640 aa Download sequence Send to blast |
MNFIFRSKPK EFAPPPPAPA TALQQQQHAA GENHRESLMM DLDLDLDASW SFDQIFAAAA 60 SASNPMSPFL VPAASEQPCS PLWAFSDENE DKPNGNALSS GSLRLSNYPR FVTYANEHEA 120 APETVSVTDD KKRIPLPIKG LAPLDYLDSS CIIKERMTQA LRYFKESTGE RVLAQIWAPV 180 KNGGRYVLTT SGQPFVLDPD CNGLHQYRMV SLMYMFSVDG ETDGVLGLPG RVYRKKLPEW 240 TPNVQYYSSK EFPRLNHALD YNVRGTLALP VFEPSGQSCV GVLELIMTSQ KINYAPEVDK 300 VCKALEAVNL KSSEILDYPN HQICNEGRQN ALVEILEILT AVCETYKLPL AQTWVPCRHR 360 SVLADGGGFK KSCSSFDGSC MGQVCMSTTD VAFYVVDAHM WGFREACAEH HLQKGQGVAG 420 RAYASQKSCF CEDIGQFCKT EYPLVHYARL FGLSSCLAIC LRSTHTGNDD YILEFFLPPN 480 DGDYTDQLAL LNSLLLTMKQ HFRSLRVASG EELEHNWGSV EIIKASTEEK LGSRFDSVPT 540 TKSLPQSASV ANGRTHPDLM EEQQSPVALN VAKGAEGVNG TAEAHNHASV PENKQTGKKS 600 ERKRGKAEKT ISLEVLQQYF AGSLKDAAKS LGGTLFKIL* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00449 | DAP | Transfer from AT4G24020 | Download |
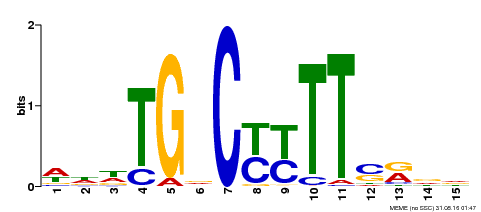
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PGSC0003DMP400021691 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG975447 | 0.0 | HG975447.1 Solanum pennellii chromosome ch08, complete genome. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006343817.1 | 0.0 | PREDICTED: protein NLP6-like | ||||
| Swissprot | Q8RWY4 | 0.0 | NLP6_ARATH; Protein NLP6 | ||||
| TrEMBL | M1AWJ9 | 0.0 | M1AWJ9_SOLTU; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400031945 | 0.0 | (Solanum tuberosum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G64530.1 | 0.0 | Nin-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | PGSC0003DMP400021691 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



