 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PGSC0003DMP400028727 | ||||||||
| Common Name | LOC102584400 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | E2F/DP | ||||||||
| Protein Properties | Length: 479aa MW: 52576.5 Da PI: 4.7344 | ||||||||
| Description | E2F/DP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | E2F_TDP | 86.3 | 2.3e-27 | 156 | 221 | 1 | 71 |
E2F_TDP 1 rkeksLrlltqkflkllekseegivtlnevakeLvsedvknkrRRiYDilNVLealnliekkekneirwkg 71
r+++sL+llt+kf++l++++e+g+++ln++a++L +v ++RRiYDi+NVLe+++liekk kn+i+wkg
PGSC0003DMP400028727 156 RYDSSLGLLTKKFINLIKHAEDGMLDLNQAADTL---EV--QKRRIYDITNVLEGIGLIEKKLKNRIQWKG 221
6899******************************...99..****************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.10 | 1.4E-29 | 154 | 223 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SuperFamily | SSF46785 | 2.54E-17 | 156 | 219 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM01372 | 1.6E-34 | 156 | 221 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| Pfam | PF02319 | 1.4E-24 | 158 | 221 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| CDD | cd14660 | 9.64E-48 | 232 | 338 | No hit | No description |
| SuperFamily | SSF144074 | 1.65E-33 | 235 | 338 | No hit | No description |
| Pfam | PF16421 | 2.0E-33 | 237 | 336 | IPR032198 | E2F transcription factor, CC-MB domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005667 | Cellular Component | transcription factor complex | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 479 aa Download sequence Send to blast |
MTGSSASTGA AAPPTAQSSS AAVNGGGRSQ ISHPIKRYLP FASMRPPFVT PEDYHRFSIP 60 GVDSRIGSAS LQPEAIVVKS PALKRKTGYI KEVESSDWTA NSGYADVANS PLGTPVSGKG 120 GRAHGRSRAT KNNKSGPQTP VYNAGSPSPL TPASCRYDSS LGLLTKKFIN LIKHAEDGML 180 DLNQAADTLE VQKRRIYDIT NVLEGIGLIE KKLKNRIQWK GVDSSRPGEV DNDASILKAE 240 TDRLSMEERR LDERIREMQE KLRDMSEDEN NQRWLFVTEE DIKSLPCFQN ETLIAVKAPH 300 GTTLEVPDPD EAVDYPQRRY RIILRSTMGP IDVYLVSQFE EKFEEMNGVE PPTTIPVASS 360 SGSKDYPTME ISTVSNSVTE NEEQTHNVDH LSSDLGTSQD YNGGMMKILP SDVDNDSDYW 420 LLSDANVSIT DMWKTDSTVE WDGETLLRDF GIDDLGTPRA HTPSTIADVP DPMNVPPR* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1cf7_A | 7e-26 | 149 | 222 | 2 | 74 | PROTEIN (TRANSCRIPTION FACTOR E2F-4) |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds DNA cooperatively with DP proteins through the E2 recognition site, 5'-TTTC[CG]CGC-3' found in the promoter region of a number of genes whose products are involved in cell cycle regulation or in DNA replication. The binding of retinoblastoma-related proteins represses transactivation. Regulates gene expression both positively and negatively. Activates the expression of E2FB. Involved in the control of cell-cycle progression from G1 to S phase. Stimulates cell proliferation and delays differentiation. {ECO:0000269|PubMed:11669580, ECO:0000269|PubMed:11786543, ECO:0000269|PubMed:11862494, ECO:0000269|PubMed:11889041, ECO:0000269|PubMed:11891240, ECO:0000269|PubMed:12913157, ECO:0000269|PubMed:15377755, ECO:0000269|PubMed:16514015, ECO:0000269|PubMed:19662336}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00661 | PBM | Transfer from Pp3c1_6880 | Download |
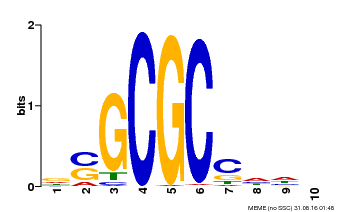
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PGSC0003DMP400028727 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG975440 | 1e-121 | HG975440.1 Solanum pennellii chromosome ch01, complete genome. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006348486.1 | 0.0 | PREDICTED: transcription factor E2FA isoform X1 | ||||
| Swissprot | Q9FNY0 | 1e-164 | E2FA_ARATH; Transcription factor E2FA | ||||
| TrEMBL | M1BCW1 | 0.0 | M1BCW1_SOLTU; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400042347 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA1077 | 24 | 84 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G36010.1 | 1e-168 | E2F transcription factor 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | PGSC0003DMP400028727 |
| Entrez Gene | 102584400 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



