 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PGSC0003DMP400032526 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 238aa MW: 26785.9 Da PI: 10.6669 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 35 | 2.6e-11 | 14 | 58 | 53 | 99 |
EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EEEEE-S CS
B3 53 rkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelvvkvfrk 99
+++++r++lt+GW+ Fv+a++L +gD+v+F ++++ +l+++++r+
PGSC0003DMP400032526 14 EGQPKRHLLTTGWSVFVSAKRLVAGDSVLFI--WNEKNQLFLGIRRA 58
678999*************************..8889999****997 PP
| |||||||
| 2 | Auxin_resp | 118.8 | 4.3e-39 | 83 | 166 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalk.vkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
aahaa+t+s F+v++nPras+seFv++++k+ ka++ ++vsvGmRf+m fete+ss rr++Gt++g+ dldpvrW nS+Wrs+k
PGSC0003DMP400032526 83 AAHAAATNSCFTVFFNPRASPSEFVIPLSKYIKAVYhTRVSVGMRFRMLFETEESSVRRYMGTITGIGDLDPVRWANSHWRSVK 166
79*********************************9789******************************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50863 | 9.925 | 1 | 59 | IPR003340 | B3 DNA binding domain |
| SuperFamily | SSF101936 | 2.35E-14 | 11 | 86 | IPR015300 | DNA-binding pseudobarrel domain |
| Pfam | PF02362 | 6.1E-9 | 14 | 58 | IPR003340 | B3 DNA binding domain |
| Gene3D | G3DSA:2.40.330.10 | 1.3E-15 | 15 | 72 | IPR015300 | DNA-binding pseudobarrel domain |
| Pfam | PF06507 | 2.0E-34 | 83 | 166 | IPR010525 | Auxin response factor |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0009908 | Biological Process | flower development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 238 aa Download sequence Send to blast |
MKLSTSGMGQ QAHEGQPKRH LLTTGWSVFV SAKRLVAGDS VLFIWNEKNQ LFLGIRRATR 60 PQTVMPSSVL SSDSMHIGLL AAAAHAAATN SCFTVFFNPR ASPSEFVIPL SKYIKAVYHT 120 RVSVGMRFRM LFETEESSVR RYMGTITGIG DLDPVRWANS HWRSVKVGWD ESTAGERQPR 180 VSLWEIEPLT TFPMYPSLFP LRLKRPWYPG TSSFQGILRT QCYAVRKVKL YNPEVSL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldu_A | 9e-74 | 13 | 187 | 214 | 388 | Auxin response factor 5 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). Seems to act as transcriptional activator. Formation of heterodimers with Aux/IAA proteins may alter their ability to modulate early auxin response genes expression. Regulates both stamen and gynoecium maturation. Promotes jasmonic acid production. Partially redundant with ARF8. {ECO:0000269|PubMed:12036261, ECO:0000269|PubMed:16107481}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00033 | PBM | Transfer from AT5G37020 | Download |
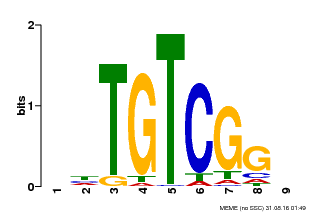
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PGSC0003DMP400032526 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by miR167. {ECO:0000269|PubMed:17021043}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HM560979 | 0.0 | HM560979.1 Solanum lycopersicum auxin response factor 8-1 (ARF8-1) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006343312.1 | 1e-142 | PREDICTED: auxin response factor 8-like isoform X1 | ||||
| Refseq | XP_006343313.1 | 1e-143 | PREDICTED: auxin response factor 8-like isoform X2 | ||||
| Refseq | XP_015162339.1 | 1e-143 | PREDICTED: auxin response factor 8-like isoform X2 | ||||
| Swissprot | Q9ZTX8 | 1e-124 | ARFF_ARATH; Auxin response factor 6 | ||||
| TrEMBL | M1BLU5 | 1e-175 | M1BLU5_SOLTU; Auxin response factor | ||||
| STRING | PGSC0003DMT400048034 | 1e-143 | (Solanum tuberosum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G30330.1 | 1e-125 | auxin response factor 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | PGSC0003DMP400032526 |



