 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PGSC0003DMP400032529 | ||||||||
| Common Name | LOC102597704 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 753aa MW: 84277.5 Da PI: 6.471 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 72.7 | 4.6e-23 | 31 | 132 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFk 83
f+k+lt sd++++g +++p++ ae+ ++ +++ ++l+ +d++ +W++++i+r++++r++lt+GW+ Fv+a++L +gD+v+F
PGSC0003DMP400032529 31 FCKTLTASDTSTHGGFSVPRRAAEKVfppldfSQTPPC-QELIARDLHDIEWKFRHIFRGQPKRHLLTTGWSVFVSAKRLVAGDSVLFI 118
99*****************************7555555.59************************************************ PP
E-SSSEE..EEEEE-S CS
B3 84 ldgrsefelvvkvfrk 99
++++ +l+++++r+
PGSC0003DMP400032529 119 --WNEKNQLFLGIRRA 132
..8889999****997 PP
| |||||||
| 2 | Auxin_resp | 116.1 | 3e-38 | 157 | 240 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalk.vkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
aahaa+t+s F+v++nPras+seFv++++k+ ka++ ++vsvGmRf+m fete+ss rr++Gt++g+ dldpvrW nS+Wrs+k
PGSC0003DMP400032529 157 AAHAAATNSCFTVFFNPRASPSEFVIPLSKYIKAVYhTRVSVGMRFRMLFETEESSVRRYMGTITGIGDLDPVRWANSHWRSVK 240
79*********************************9789******************************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 5.89E-49 | 15 | 160 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 8.5E-41 | 24 | 146 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.17E-21 | 30 | 126 | No hit | No description |
| Pfam | PF02362 | 2.0E-21 | 31 | 132 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 2.8E-23 | 31 | 133 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 11.984 | 31 | 133 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 1.4E-33 | 157 | 240 | IPR010525 | Auxin response factor |
| PROSITE profile | PS51745 | 23.173 | 625 | 709 | IPR000270 | PB1 domain |
| SuperFamily | SSF54277 | 4.25E-7 | 627 | 699 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0009908 | Biological Process | flower development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 753 aa Download sequence Send to blast |
MTLQPLTPQE QKDTYLPVEL GIPSRQPTNY FCKTLTASDT STHGGFSVPR RAAEKVFPPL 60 DFSQTPPCQE LIARDLHDIE WKFRHIFRGQ PKRHLLTTGW SVFVSAKRLV AGDSVLFIWN 120 EKNQLFLGIR RATRPQTVMP SSVLSSDSMH IGLLAAAAHA AATNSCFTVF FNPRASPSEF 180 VIPLSKYIKA VYHTRVSVGM RFRMLFETEE SSVRRYMGTI TGIGDLDPVR WANSHWRSVK 240 VGWDESTAGE RQPRVSLWEI EPLTTFPMYP SLFPLRLKRP WYPGTSSFQE NNSEAINGMA 300 WLRGESSEQG PHLLNLQSFG GMLPWMQQRV DPTMLRNDLN QQYQAMLASG LQNFGSGDLM 360 KQQLMQFPQP VQYVQHAGSL NPLLQQQQQQ QAMQQTIHQH MLPAQTQDNL QRQQQQHVSN 420 QTEEQSHHHS YQEAYQIPNS QLQQKQPSNV PSPSFSKPDI ADPSSKFSAS IAPSGMPTAL 480 GSLCSEGTSN FLNFNILGQQ PVIMEQQQQQ QQQQQKSWMA KFAHSQLNTG SNSPSLSGYG 540 KDTSNSQETC SLDAQNQSLF GANVDSSGLL LPTTVSNVAT TSIDADISSM PLGTSGFSNS 600 LYGYVQDSSD MLHNVGQVDA QTAPRTFVKV YKSASLGRSL DITRFNSYHE LRQELGQMFG 660 IEGFLEDPQR SGWQLVFVDR ENDVLLLGDD PWEEFVNNVW YIKILSPEDV QKLGKEEVGS 720 LNRGLPERMS SNNSADGRDF MSGLSSIGSL EY* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldu_A | 1e-112 | 1 | 261 | 128 | 388 | Auxin response factor 5 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). Seems to act as transcriptional activator. Formation of heterodimers with Aux/IAA proteins may alter their ability to modulate early auxin response genes expression. Regulates both stamen and gynoecium maturation. Promotes jasmonic acid production. Partially redundant with ARF6. Involved in fruit initiation. Acts as an inhibitor to stop further carpel development in the absence of fertilization and the generation of signals required to initiate fruit and seed development. {ECO:0000269|PubMed:12036261, ECO:0000269|PubMed:16107481, ECO:0000269|PubMed:16829592}. | |||||
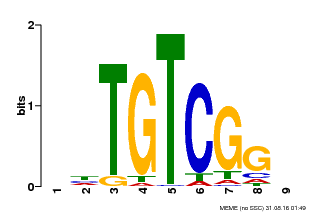
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00033 | PBM | Transfer from AT5G37020 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PGSC0003DMP400032529 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by miR167. {ECO:0000269|PubMed:17021043}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HM560979 | 0.0 | HM560979.1 Solanum lycopersicum auxin response factor 8-1 (ARF8-1) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006343312.1 | 0.0 | PREDICTED: auxin response factor 8-like isoform X1 | ||||
| Swissprot | Q9FGV1 | 0.0 | ARFH_ARATH; Auxin response factor 8 | ||||
| TrEMBL | M1BLU8 | 0.0 | M1BLU8_SOLTU; Auxin response factor | ||||
| STRING | PGSC0003DMT400048034 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3857 | 23 | 39 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G37020.1 | 0.0 | auxin response factor 8 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | PGSC0003DMP400032529 |
| Entrez Gene | 102597704 |



