 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PGSC0003DMP400037187 | ||||||||
| Common Name | LOC102602066 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 477aa MW: 52574.5 Da PI: 6.4276 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 51.7 | 2e-16 | 57 | 101 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r rW++eE++++++a k++G W++I +++g ++t+ q++s+ qk+
PGSC0003DMP400037187 57 RERWSEEEHKKFLEALKLHGRA-WRRIEEHVG-TKTAVQIRSHAQKF 101
78******************88.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 5.08E-16 | 51 | 105 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 21.204 | 52 | 106 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 2.9E-15 | 55 | 104 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 5.5E-13 | 56 | 104 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.2E-9 | 57 | 97 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 6.8E-14 | 57 | 100 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.15E-10 | 59 | 102 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 477 aa Download sequence Send to blast |
MTIGDQSESD TILPSRNRMS VDVGAPSPRS IQMEEQVIPG EEYAPKIRKP YTISKQRERW 60 SEEEHKKFLE ALKLHGRAWR RIEEHVGTKT AVQIRSHAQK FFSKVVRESS NGDASSVKSI 120 EIPPPRPKRK PMHPYPRKMV TPLKSGTLAS EKLNRSGSPD LYLSEPENQS PTSVLSAFGS 180 DAFGTVDSTK PSEQSSPVSS AVAENSGDLV LSEPSDFIVE ESRSSPARAY TSSNPANQTC 240 VKLELFPEDN DFVKEGSDEA SSTQCLKLFG KTVLVTDTYM PSSTSGQISL TDENDEPASR 300 TLPRSTFPAK YLHWDSECPR STFSIVPPTP SYYFPTPNVS PGTNQSDSTP LLPWGSSAAV 360 RCTQVHNPIP MKGRPLFNDR DLEGKETQKE GSSTGSNTES VDAELSGDKN LEIEAQSSRN 420 LVEESVRGGS LFERRANSTK RVKGFVPYKR CLAERGMSSS TLTGEEREEQ RTRLCL* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00515 | DAP | Transfer from AT5G17300 | Download |
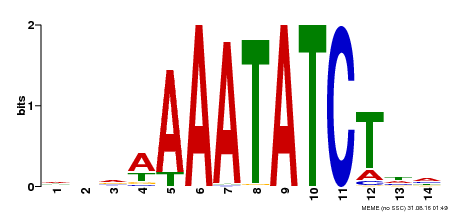
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PGSC0003DMP400037187 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | DQ103498 | 0.0 | DQ103498.1 Solanum ochranthum putative At5g37260 (CT251) gene, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006350462.1 | 0.0 | PREDICTED: protein REVEILLE 1 | ||||
| TrEMBL | M1BXP3 | 0.0 | M1BXP3_SOLTU; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400055263 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3185 | 24 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G17300.1 | 8e-68 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | PGSC0003DMP400037187 |
| Entrez Gene | 102602066 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



