 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PGSC0003DMP400044419 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 467aa MW: 50756.5 Da PI: 5.5401 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 48.2 | 2.6e-15 | 63 | 107 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r +WT+eE++++++a k++G W+ I +++g +t+ q++s+ qk+
PGSC0003DMP400044419 63 REKWTEEEHQRFLEALKLYGRA-WRQIEEYVG-SKTAIQIRSHAQKF 107
789*****************88.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 6.65E-16 | 57 | 112 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 20.982 | 58 | 112 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 3.1E-17 | 61 | 110 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 3.6E-12 | 62 | 110 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.7E-12 | 63 | 106 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-8 | 63 | 103 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 3.88E-9 | 65 | 108 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 467 aa Download sequence Send to blast |
MVADAVYAFF TIPQNQVEGA QQGASSMISS KLETVATQGK APTCLANENV LKARKPYTIT 60 KQREKWTEEE HQRFLEALKL YGRAWRQIEE YVGSKTAIQI RSHAQKFFAK IARDSGNDGD 120 ESLNAIDIPP PRPKKKPLHP YPRKIADSSV ANKAVSGQPE RSPSPNASGR DSCSPDSVLP 180 AIGLGASEYS AAEQQNSRFS PVSCTTDAHT ANVISAENDD ESMSSNSNTV EEIHVALKPV 240 AASTCPITNS EFMECDIVHM ENSCNGEKLA VEPPSASIKL FGKTVFVPDA NNLAPPALYN 300 TEKEIKISNE DVLHAFQANQ ANSPFILAMV PGNMIPPASW LSQNKLENNP ESTAAFPAAT 360 ISWWSWYQDL IYRSISSCGQ TAMETAAHYQ RPKDEESQRE GSSTGSSIGS ASEVDDGNRS 420 SETVESKCTT KSKGFVPYKR CLAERDGKPA GAVLEERESQ RVRVCS* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00146 | DAP | Transfer from AT1G18330 | Download |
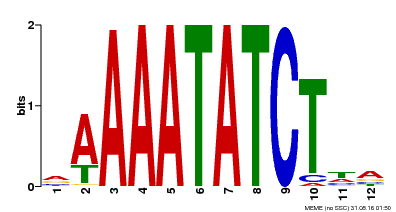
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PGSC0003DMP400044419 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC151804 | 0.0 | AC151804.1 Solanum demissum chromosome 5 clone PGEC475B22, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015071307.1 | 0.0 | uncharacterized protein LOC107015511 isoform X1 | ||||
| TrEMBL | M1CEP7 | 0.0 | M1CEP7_SOLTU; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400065850 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA7447 | 21 | 27 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G17300.1 | 2e-36 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | PGSC0003DMP400044419 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



