 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PGSC0003DMP400046284 | ||||||||
| Common Name | LOC102604336 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 328aa MW: 36713.2 Da PI: 8.7877 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 100.1 | 1.5e-31 | 64 | 117 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
+rlrWtpeLH rFv+ave+LGG+++AtPk +l++m++kgL+++hvkSHLQ++R+
PGSC0003DMP400046284 64 ARLRWTPELHLRFVHAVERLGGQDRATPKLVLQMMNIKGLSIAHVKSHLQMFRS 117
69***************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 12.661 | 60 | 120 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.5E-29 | 60 | 118 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 5.91E-15 | 63 | 118 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.6E-22 | 64 | 119 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.1E-9 | 65 | 115 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 328 aa Download sequence Send to blast |
MLEDSGETGC NSKTSSNFEK NEDEVLEENA SSKFKDGGSS SESTLEESEK IKPCVRPYVR 60 SKMARLRWTP ELHLRFVHAV ERLGGQDRAT PKLVLQMMNI KGLSIAHVKS HLQMFRSKKI 120 DDQSQGIGHH HKLFMEGGDP NIFNMIQFPR FPAYHQRLNS TFRYGDASWN CHGNWMPSNT 180 MGQRIPSFIN ERSNLQRNEI KDVMGSSIGS PSGELTRLFH IASKAQARAF VGNGITSPPN 240 LERAITPLKR KVSYNDQVDL NLYLGVKPRN EVTPNLDDND NDNISSLSLS LSSPLSYSRA 300 TRFIEDANID GKTENARRGA STLDLTL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 3e-18 | 64 | 119 | 2 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 3e-18 | 64 | 119 | 2 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 3e-18 | 64 | 119 | 2 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 3e-18 | 64 | 119 | 2 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00297 | DAP | Transfer from AT2G38300 | Download |
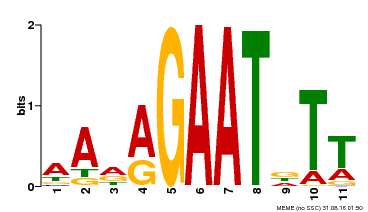
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PGSC0003DMP400046284 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG975448 | 0.0 | HG975448.1 Solanum pennellii chromosome ch09, complete genome. | |||
| GenBank | HG975521 | 0.0 | HG975521.1 Solanum lycopersicum chromosome ch09, complete genome. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006359562.2 | 0.0 | PREDICTED: uncharacterized protein LOC102604336 | ||||
| TrEMBL | M1CJ60 | 0.0 | M1CJ60_SOLTU; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400068508 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4382 | 21 | 41 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38300.1 | 4e-51 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | PGSC0003DMP400046284 |
| Entrez Gene | 102604336 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



