 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PGSC0003DMP400048281 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 784aa MW: 85755 Da PI: 5.5593 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 65.9 | 5.4e-21 | 126 | 181 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe+lF+++++p++++r eL+k+l+L++rqVk+WFqNrR+++k
PGSC0003DMP400048281 126 KKRYHRHTPQQIQELESLFKECPHPDEKQRLELSKRLSLETRQVKFWFQNRRTQMK 181
688999***********************************************999 PP
| |||||||
| 2 | START | 198.2 | 3.6e-62 | 333 | 557 | 1 | 206 |
HHHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S... CS
START 1 elaeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdetla... 77
ela++a++elvk+a+ +ep+W++s e +n +e++++f++ + + +ea+r++g+v+ ++ lve+l+d++ +W e+++
PGSC0003DMP400048281 333 ELALAAMEELVKLAQTDEPLWFRSIeggrELLNHEEYIRTFTPCIGmrpnsFVSEASRETGMVIINSLALVETLMDSN-KWAEMFPcli 420
5899***********************9999***********9999****9***************************.********** PP
.EEEEEEEECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEEC CS
START 78 .kaetlevissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepks 158
+ +t++vissg galqlm aelq+lsplvp R++ f+R+++q+ +g+w++vdvS+d ++ + + + +++lpSg+++++++
PGSC0003DMP400048281 421 aRTSTTDVISSGmggtrnGALQLMHAELQVLSPLVPiREVNFLRFCKQHAEGVWAVVDVSIDTIRETSGAPTYPNCRRLPSGCVVQDMP 509
******************************************************************999******************** PP
TCEEEEEEEE-EE--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 159 nghskvtwvehvdlkgrlphwllrslvksglaegaktwvatlqrqcek 206
ng+skvtwveh+++++ h l+r+l++ g+ +ga++wvatlqrqce+
PGSC0003DMP400048281 510 NGYSKVTWVEHAEYEEGANHHLYRQLISAGMGFGAQRWVATLQRQCEC 557
**********************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 3.4E-21 | 112 | 177 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.71E-20 | 113 | 183 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.637 | 123 | 183 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 5.2E-19 | 124 | 187 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 4.91E-19 | 125 | 183 | No hit | No description |
| Pfam | PF00046 | 1.6E-18 | 126 | 181 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 158 | 181 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 44.059 | 324 | 560 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.28E-31 | 326 | 557 | No hit | No description |
| CDD | cd08875 | 3.51E-123 | 328 | 556 | No hit | No description |
| SMART | SM00234 | 3.4E-47 | 333 | 557 | IPR002913 | START domain |
| Pfam | PF01852 | 6.2E-54 | 333 | 557 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 6.41E-21 | 576 | 776 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 784 aa Download sequence Send to blast |
MNFGGFLDNN SGGGGARIVA DIPFNHNSSS NNDNKNNNMP TGAISQPRLL PQSLAKNMFN 60 SPGLSLALQT GMEGQNEVTR MAENYEGNNS VGRRSREEEP DSRSGSDNLE GASGDEQDAA 120 DKPPRKKRYH RHTPQQIQEL ESLFKECPHP DEKQRLELSK RLSLETRQVK FWFQNRRTQM 180 KTQLERHENS ILRQENDKLR AENMSIREAM RNPICTNCGG PAMIGEISLE EQHLRIENAR 240 LKDELDRVCA LAGKFLGRPI SSLVTSMPPP MPNSSLELGV GNNGYGGMSN VPTTLPLAPP 300 DFGVGISNSL PVVPSNRQST GIERSLERSM YLELALAAME ELVKLAQTDE PLWFRSIEGG 360 RELLNHEEYI RTFTPCIGMR PNSFVSEASR ETGMVIINSL ALVETLMDSN KWAEMFPCLI 420 ARTSTTDVIS SGMGGTRNGA LQLMHAELQV LSPLVPIREV NFLRFCKQHA EGVWAVVDVS 480 IDTIRETSGA PTYPNCRRLP SGCVVQDMPN GYSKVTWVEH AEYEEGANHH LYRQLISAGM 540 GFGAQRWVAT LQRQCECLAI LMSSTVSARD HTGNVDEDVR VMTRKSVDDP GEPAGIVLSA 600 ATSVWLPVSP QRLFDFLRDE RLRSEWDILS NGGPMQEMAH IAKGQDHGNC VSLLRASAMN 660 ANQSSMLILQ ETCIDAAGAL VVYAPVDIPA MHVVMNGGDS AYVALLPSGF SIVPDGPGSR 720 GSNGPSCNGG PDQRISGSLL TVAFQILVNS LPTAKLTVES VETVNNLISC TVQKIKAALQ 780 CES* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of the tissue-specific accumulation of anthocyanins and in cellular organization of the primary root. {ECO:0000269|PubMed:10402424}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
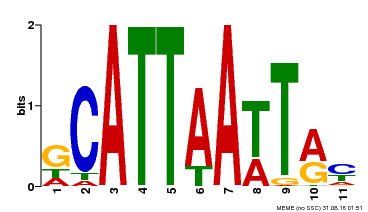
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PGSC0003DMP400048281 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | GQ222185 | 0.0 | GQ222185.1 Solanum lycopersicum cultivar M82 cutin deficient 2 (CD2) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006339457.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| Swissprot | Q0WV12 | 0.0 | ANL2_ARATH; Homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| TrEMBL | M1CNN2 | 0.0 | M1CNN2_SOLTU; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400071377 | 0.0 | (Solanum tuberosum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00730.1 | 0.0 | HD-ZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | PGSC0003DMP400048281 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



