 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PGSC0003DMP400048856 | ||||||||
| Common Name | LOC102600966 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 348aa MW: 37872.5 Da PI: 8.7664 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 49.9 | 6.8e-16 | 277 | 319 | 5 | 47 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkk 47
+r++r++kNRe+A rsR+RK+a++ eLe+kv Le+eN++Lk
PGSC0003DMP400048856 277 RRQKRMIKNRESAARSRARKQAYTHELENKVSRLEEENERLKR 319
79*************************************9993 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 8.5E-16 | 268 | 320 | No hit | No description |
| SMART | SM00338 | 1.8E-14 | 273 | 345 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 11.634 | 275 | 320 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 5.9E-13 | 277 | 320 | IPR004827 | Basic-leucine zipper domain |
| CDD | cd14707 | 1.06E-24 | 277 | 322 | No hit | No description |
| SuperFamily | SSF57959 | 3.13E-11 | 277 | 322 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 280 | 295 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 348 aa Download sequence Send to blast |
MGSQGGGGGG GGGVGVNSIG ATQTQAQAQA HAQDPKTNAL ARQGSLYSLT LDEVQNQLGD 60 LGKPLSSMNL DELLKTVWTV EASQGMGGTD YGVLQHGQDA SGSSLNRQSS ITLTSDLSKK 120 TVDQVWQDIQ QGHKRDSIDR KAQERQPTLG EMTLEDFLVK AGVVAESTPG KKSSGSVLGV 180 DSMALPQQQA QWSQYQMHAM HQLPPQQQQQ NMLPVFMPGH SVQQPLTIVS NPTIDAAYPE 240 SQMTMSPTAL LGTLSDTQTL GRKRVAPDDV VEKTVERRQK RMIKNRESAA RSRARKQAYT 300 HELENKVSRL EEENERLKRQ KEIEQVLPSV PLPEPKYQLR RTSSAPV* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds to the embryo specification element and the ABA-responsive element (ABRE) of the Dc3 gene promoter. Could participate in abscisic acid-regulated gene expression during seed development. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00409 | DAP | Transfer from AT3G56850 | Download |
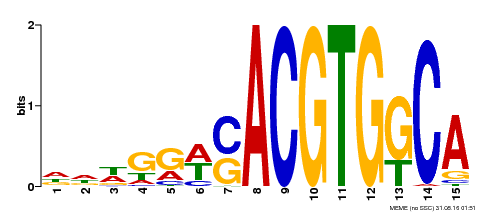
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PGSC0003DMP400048856 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG975522 | 0.0 | HG975522.1 Solanum lycopersicum chromosome ch10, complete genome. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006339213.1 | 0.0 | PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 2 | ||||
| Swissprot | Q9LES3 | 1e-100 | AI5L2_ARATH; ABSCISIC ACID-INSENSITIVE 5-like protein 2 | ||||
| TrEMBL | M1CPY9 | 0.0 | M1CPY9_SOLTU; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400072260 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3334 | 24 | 48 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G56850.1 | 2e-99 | ABA-responsive element binding protein 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | PGSC0003DMP400048856 |
| Entrez Gene | 102600966 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



