 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PGSC0003DMP400051154 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 255aa MW: 27474.5 Da PI: 10.4798 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 30.8 | 5.9e-10 | 234 | 254 | 2 | 22 |
--SS-EEEEEEE--TT-SS-E CS
WRKY 2 dDgynWrKYGqKevkgsefpr 22
D+++WrKYGqK++kgs +pr
PGSC0003DMP400051154 234 ADEFSWRKYGQKPIKGSAYPR 254
69******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF10533 | 4.1E-13 | 185 | 231 | IPR018872 | Zn-cluster domain |
| Gene3D | G3DSA:2.20.25.80 | 7.7E-9 | 221 | 254 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 11.13 | 228 | 254 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 9.68E-7 | 231 | 254 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.2E-5 | 235 | 254 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:1990841 | Molecular Function | promoter-specific chromatin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 255 aa Download sequence Send to blast |
MAVVSKMTEN FAVQEAASAG LKSMEHLIRL VSHEPVQVDC REIADFTVSK FKKVISILDR 60 TGHARFRRGP VQAPVPVYPD SFTTLSLAPS LSFAPAKETP AVQLQTALTL DFTKPSVERP 120 IGNSSAFTAF AVKSKEVLMA TPTPTNSSSF MSSITGEATV SNGKQASSSM LLLPAQAGNL 180 PITGKRCREH EQSDDISGSK STGSGKCHCK KRKSRDKKVM RIAAISSRIA DIPADEFSWR 240 KYGQKPIKGS AYPR* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). Regulates rhizobacterium B.cereus AR156-induced systemic resistance (ISR) to P.syringae pv. tomato DC3000, probably by activating the jasmonic acid (JA)- signaling pathway (PubMed:26433201). {ECO:0000250, ECO:0000269|PubMed:26433201}. | |||||
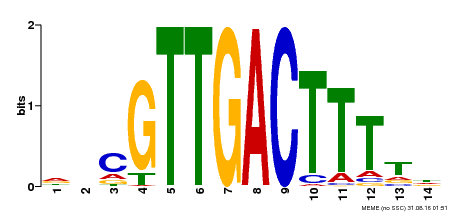
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00464 | DAP | Transfer from AT4G31550 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PGSC0003DMP400051154 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Strongly during leaf senescence (PubMed:11722756). Repressed by rhizobacterium B.cereus AR156 in leaves, and to a lower extent, by P.fluorescens WCS417r (PubMed:26433201). {ECO:0000269|PubMed:11722756, ECO:0000269|PubMed:26433201}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT014501 | 0.0 | BT014501.1 Lycopersicon esculentum clone 133872F, mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006342466.1 | 0.0 | PREDICTED: probable WRKY transcription factor 17 | ||||
| Swissprot | Q9SV15 | 1e-66 | WRK11_ARATH; Probable WRKY transcription factor 11 | ||||
| TrEMBL | M1CVB8 | 0.0 | M1CVB8_SOLTU; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400075531 | 0.0 | (Solanum tuberosum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G31550.2 | 6e-60 | WRKY DNA-binding protein 11 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | PGSC0003DMP400051154 |



