 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PGSC0003DMP400055717 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Solaneae; Solanum
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 132aa MW: 14822 Da PI: 9.1689 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 76.6 | 3.9e-24 | 59 | 107 | 2 | 50 |
-SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEE CS
SBP 2 CqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfh 50
Cq+e+C++dls+ak+yh+rhkvCe h+ka+vv+v+gl+qrfCqqCsr
PGSC0003DMP400055717 59 CQAEKCNVDLSDAKQYHKRHKVCEYHAKAQVVVVAGLRQRFCQQCSRLD 107
***********************************************74 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 1.1E-23 | 53 | 107 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 17.349 | 56 | 131 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 4.58E-22 | 58 | 108 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 2.1E-18 | 59 | 107 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010321 | Biological Process | regulation of vegetative phase change | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 132 aa Download sequence Send to blast |
METANNNQLK SMEKNDDFLV ISTSENAKKK IITTNNNNNK KLSSSSNSSS SSSSLIRSCQ 60 AEKCNVDLSD AKQYHKRHKV CEYHAKAQVV VVAGLRQRFC QQCSRLDHYI SLCLVLCDTL 120 IFHLTCLKHK L* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 5e-19 | 49 | 106 | 1 | 58 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00634 | PBM | Transfer from PK22320.1 | Download |
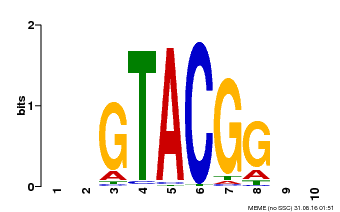
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PGSC0003DMP400055717 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG975446 | 1e-114 | HG975446.1 Solanum pennellii chromosome ch07, complete genome. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006367482.1 | 4e-69 | PREDICTED: squamosa promoter-binding protein 1-like | ||||
| TrEMBL | M1D692 | 9e-89 | M1D692_SOLTU; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400082789 | 1e-68 | (Solanum tuberosum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G02065.2 | 5e-23 | squamosa promoter binding protein-like 8 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | PGSC0003DMP400055717 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



