 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PH01000019G1890 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Bambusoideae; Arundinarodae; Arundinarieae; Arundinariinae; Phyllostachys
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 378aa MW: 43469.5 Da PI: 8.3445 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 17.5 | 1.1e-05 | 70 | 91 | 2 | 23 |
EETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 2 kCpdCgksFsrksnLkrHirtH 23
+C+ Cg sF + +Lk+H+++H
PH01000019G1890 70 TCQECGSSFQKPAHLKQHMQSH 91
6*******************99 PP
| |||||||
| 2 | zf-C2H2 | 23.1 | 1.9e-07 | 97 | 121 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
+ Cp dC+ s++rk++L+rH+ +H
PH01000019G1890 97 FICPleDCPFSYRRKDHLNRHMLKH 121
78*********************99 PP
| |||||||
| 3 | zf-C2H2 | 14.4 | 0.00011 | 126 | 151 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
++Cp C++ Fs k n +rH + H
PH01000019G1890 126 FTCPmdGCDRKFSIKANIQRHVKEiH 151
89********************9988 PP
| |||||||
| 4 | zf-C2H2 | 15.9 | 3.8e-05 | 164 | 187 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirtH 23
+C+ C+k F+ s Lk+H +H
PH01000019G1890 164 VCNeeGCNKAFKYASKLKKHEESH 187
588889**************9988 PP
| |||||||
| 5 | zf-C2H2 | 13.1 | 0.00028 | 254 | 278 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirt.H 23
kC+ C sFs+ksnL++H + H
PH01000019G1890 254 KCNfeGCECSFSNKSNLTKHTKAcH 278
79999****************9855 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00355 | 55 | 23 | 46 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 5.1E-9 | 66 | 89 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 14.274 | 69 | 96 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.014 | 69 | 91 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 71 | 91 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 3.56E-12 | 82 | 123 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 9.4E-14 | 90 | 123 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 9.099 | 97 | 126 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 2.2E-4 | 97 | 121 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 99 | 121 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 2.3E-7 | 124 | 152 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 8.29E-7 | 124 | 152 | No hit | No description |
| SMART | SM00355 | 9.0E-4 | 126 | 151 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.055 | 126 | 156 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 128 | 151 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.088 | 163 | 187 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 9.452 | 163 | 187 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 4.2E-6 | 164 | 187 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 165 | 187 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 16 | 195 | 220 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 197 | 220 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 11 | 223 | 244 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 7.53E-7 | 236 | 295 | No hit | No description |
| PROSITE profile | PS50157 | 11.302 | 253 | 283 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.68 | 253 | 278 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 4.5E-5 | 254 | 278 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 255 | 278 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 4.0E-4 | 279 | 306 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 3.1 | 284 | 310 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 286 | 310 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 378 aa Download sequence Send to blast |
MSGDEIDGDA RVEETRCRDI RRYKCEFCTV IRSKKCLIRS HMVDHHKDEL DKSDIYKSNG 60 EKVVHERENT CQECGSSFQK PAHLKQHMQS HSDEKPFICP LEDCPFSYRR KDHLNRHMLK 120 HQGKLFTCPM DGCDRKFSIK ANIQRHVKEI HEDGNVTKSG QLAVCNEEGC NKAFKYASKL 180 KKHEESHVKL DYVEVVCCEP GCMKTLTNVE CLKAHTLSCH QYVQCDICGK KHLKKNIKRH 240 LRAHDVVPST ERMKCNFEGC ECSFSNKSNL TKHTKACHDQ AKPFACRFVG CGKVFPYKHV 300 RDNHEKSSAH VYVEGDFEEM DEQLRSRPRG GRKRKAVTVE TLTRKRVTMP CEASSLDDGA 360 EYMRWLLSGG DDSSHTQ* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1tf6_A | 1e-18 | 73 | 278 | 19 | 188 | PROTEIN (TRANSCRIPTION FACTOR IIIA) |
| 1tf6_D | 1e-18 | 73 | 278 | 19 | 188 | PROTEIN (TRANSCRIPTION FACTOR IIIA) |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 326 | 334 | RPRGGRKRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Essential protein (PubMed:22353599). Isoform 1 is a transcription activator the binds both 5S rDNA and 5S rRNA and stimulates the transcription of 5S rRNA gene (PubMed:12711688, PubMed:22353599). Isoform 1 regulates 5S rRNA levels during development (PubMed:22353599). {ECO:0000269|PubMed:12711688, ECO:0000269|PubMed:22353599}. | |||||
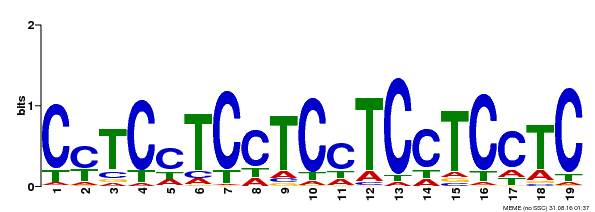
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PH01000019G1890 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015626855.1 | 0.0 | transcription factor IIIA isoform X1 | ||||
| Swissprot | Q84MZ4 | 1e-121 | TF3A_ARATH; Transcription factor IIIA | ||||
| TrEMBL | A0A0E0CDW2 | 0.0 | A0A0E0CDW2_9ORYZ; Uncharacterized protein | ||||
| STRING | OMERI02G00620.1 | 0.0 | (Oryza meridionalis) | ||||
| STRING | ORGLA02G0010700.1 | 0.0 | (Oryza glaberrima) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2382 | 38 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.2 | 1e-123 | transcription factor IIIA | ||||



