 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | PH01000040G1230 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Bambusoideae; Arundinarodae; Arundinarieae; Arundinariinae; Phyllostachys
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 950aa MW: 103584 Da PI: 5.9523 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 131.3 | 3.3e-41 | 136 | 213 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+CqvegC adl+ ak+yhrrhkvCe+h+ka++++v ++ qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+++
PH01000040G1230 136 ACQVEGCGADLTAAKDYHRRHKVCEMHAKATTAVVGNVVQRFCQQCSRFHLLQEFDEGKRSCRRRLAGHNRRRRKARP 213
5**************************************************************************875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 5.8E-34 | 130 | 198 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.624 | 134 | 211 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.29E-38 | 135 | 215 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 2.9E-30 | 137 | 210 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF48403 | 5.33E-7 | 708 | 844 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 8.667 | 741 | 804 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 3.2E-7 | 742 | 843 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 4.63E-8 | 746 | 842 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 950 aa Download sequence Send to blast |
MEAARVGAQS RQKRVFGWDL NDWSWDSERF VATPVLAAAG NGLALNSSPS SSEEAEAQVA 60 RSGNVRGDSD KRKRVVVIDD DDVEDEDPMD SGGGALSLRI GGDGVSGGAV EGGGVNEEER 120 NGKKIRVQGD SSSGPACQVE GCGADLTAAK DYHRRHKVCE MHAKATTAVV GNVVQRFCQQ 180 CSRFHLLQEF DEGKRSCRRR LAGHNRRRRK ARPEIAVGGT ASIEDKVGSY LLLSLLGICA 240 NLNSDNAEHL KGHELLSNLW RNLGTVAKSL DPKELCKLLE ACQSMQNGSN AGTSETANAL 300 FNTAAAEAAG PSNSKVPFVN GGRCGQASSS VAPVQSKATM VATPEPPACK FKDFDLNNTC 360 IDMEGFEDGY KGSPAPAFKA TDSPNCPSWM QQDSTQSPPQ TSGNSDSTSA QSLSSSNGDA 420 QYRTDKIVFK LFEKVPSDLP PVLRSQILGW LSSSPTDIES YIRPGCIILT VYVRLVESAW 480 RELSDNMSSH LVKLLSSSSD NFWASGLVFV MVRHQIAFMH NGQVTLDRPL APNSHRYCKI 540 LCVQPVAVPY SATVNFKVEG FNLVGASSRL ICSFEGRCIF QEDTAIVADD VEHEDTECLN 600 FCCPLPGSRG RGFIEAQLVE DGGFSNGFFP FIIAEQDVCT EVCELESIFK SSSYEQADDD 660 NSRNQALEFL NELGWLLHRA NTISKHDKVE LPQAAFNLWR FRNLGIFAME REWCAVTKML 720 LDFLFIGLVD VGSQSPEEVV LSENLLHTAV QRKSVRMVRF LLRYKPNKSL NTYLFRPDAW 780 GPSTITPLHI AAATTDAEDV LDALTDDPGL VGISVWRNAR DETGFTPEDY ARQRGNDAYM 840 NMVQKKIDKH LGKGHVVLGV PSNMCPVITD GVKPGGMPLA PPVPSCNICS RQALMYPNSA 900 ARTFLYRPAM LTVMGVAVIC VCVGILLHTF PKVYAAPTFR WELLERGAM* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 2e-30 | 131 | 210 | 5 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00633 | PBM | Transfer from PK06791.1 | Download |
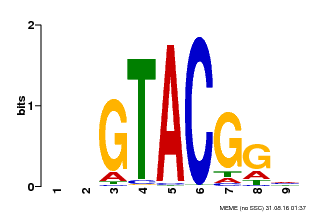
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | PH01000040G1230 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015631511.1 | 0.0 | squamosa promoter-binding-like protein 6 isoform X1 | ||||
| Swissprot | Q75LH6 | 0.0 | SPL6_ORYSJ; Squamosa promoter-binding-like protein 6 | ||||
| TrEMBL | A0A3Q8ANT3 | 0.0 | A0A3Q8ANT3_9POAL; Squamosa-promoter binding protein-like protein | ||||
| STRING | OGLUM03G39810.1 | 0.0 | (Oryza glumipatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1998 | 38 | 92 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47070.1 | 0.0 | squamosa promoter binding protein-like 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



